Introduction
mRNA therapeutics has demonstrated significant potential, paving the way for next-generation treatments across various fields1,2,3, including infectious diesease4,5,6,7,8, cancer vaccines9,10, protein replacement11,12, gene editing13,14,15, and immune therapies16,17,18,19,20. Despite its promise, the mRNA platform faces obstacles such as high costs for modified bases and a short in vivo half-life, which hinder further advancements21,22. To overcome these challenges, exploring alternative RNA platforms like circRNA offers a promising direction. CircRNA has demonstrated promise in the development of vaccines for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), achieving effectiveness without the need for base modifications, thereby establishing itself as a compelling candidate for next-generation RNA therapeutics23.
Discovered in 1976, circRNA is a covalently closed, single-stranded RNA ring molecule that lacks free 5’ and 3’ ends. It has been found across numerous species24,25, displaying increased stability and an extended half-life compared to linear RNA26,27. In vitro-transcribed (IVT) circRNAs similarly exhibit improved stability23,28,29. Lacking a canonical 5’ cap structure, circRNAs can initiate translation through internal ribosomal entry site (IRES) or IRES-like elements28,30,31, enabling protein expression in both cells and animals23,28,29,32. This translational capacity has opened the door to numerous promising therapeutic applications23,33,34,35,36,37. In addition to coding functions, non-coding circRNAs also play crucial roles in RNA-based therapeutics38,39,40,41. Recently, circRNA-based therapeutics have progressed into clinical trials, with candidates such as RXRG001 (RiboX Therapeutics), HM2002 (CircCode Biomed), and TI-0083 (Therorna Inc.) representing the field’s translational momentum.
In vitro RNA circularization techniques are fundamental to the circRNA platform, with methods including chemical synthesis and circularization42,43,44,45, ligases-mediated23,46,47, and ribozymes-mediated circularization28,48,49,50,51, each with distinct limitations52,53. Among these, self-splicing ribozymes, particularly from group I and group II introns, are frequently used due to their efficiency and simplicity, as they bypass the need for proteins. For instance, the Anabaena (Ana) group I intron has been engineered to produce circRNAs using permuted intron-exon (PIE)28,39,54,55, which is widely applied in circRNA-based theraputics23,33,34,38,39,56,57. PIE methods have shown potential for adaptation using alternative introns58 or different intron split sites59; however, achieving high circularization efficiency remains challenging, and only a limited number of introns are compatible with this approach. Group II introns also hold promise—particularly for producing circRNAs without exogenous sequences60 or with reduced levels of nicked RNA byproducts61. Additionally, the trans-splicing activity of group I introns has been applied to RNA circularization49,50,51, with high efficiency observed in several specific introns62.
However, current RNA circularization platforms still face several limitations. The PIE method, for instance, relies on specific intron split sites, limiting its adaptability and efficiency when applied to other introns28,58,63. Efficient generation of large circRNAs—particularly those exceeding 9-kb—also remains a significant technical hurdle28,39,51. Additionally, the immunogenicity of in vitro-synthesized circRNA, likely due to impurities or foreign sequences (scars)29,64, calls for scarless, high-purity circRNA production.
In this study, we report two RNA circularization techniques: Permuted Intron-Exon through Trans-splicing (PIET) and Complete self-splicing Intron for RNA Circularization (CIRC). PIET offers an alternative RNA circularization method, while CIRC stands out by effectively leveraging intact group I and group II introns, opening avenues to identify superior introns. CIRC surpasses PIE by increasing circularization efficiency and reducing the need for high concentration of Mg2+, high pH, and lengthy incubation times, supporting rapid RNA circularization and maintaining RNA integrity. With these capabilities, CIRC successfully circularizes RNA encoding full-length human dystrophin (about 12,000 nucleotides), enabling the expression of the 427-kDa protein. Additionally, CIRC enables efficient scarless circRNA production and streamlines purification using RNase R or oligo(dT)-based methods. These innovations broaden the potential of circRNA platform, enhancing its applicability in RNA therapeutics.
Results
PIE can begin with the second step of the transesterification process
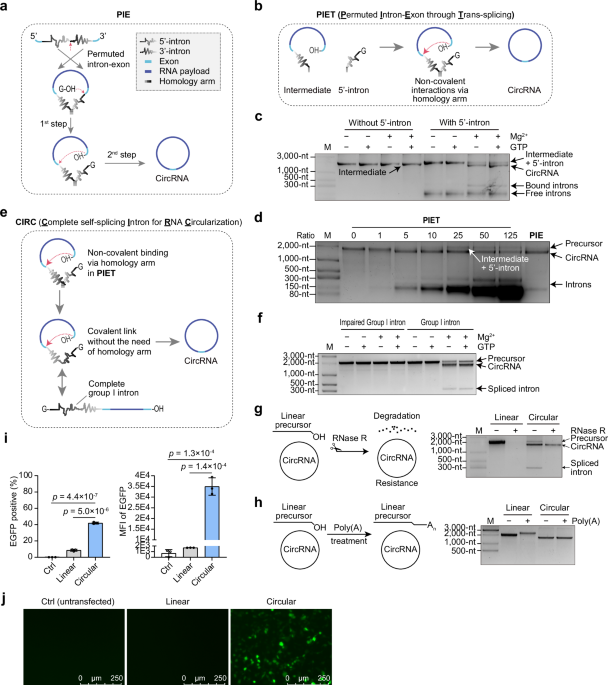
Group I introns, which are self-splicing ribozymes, enable RNA splicing without protein assistance, functioning through a two-step transesterification reaction that requires guanosine as a cofactor65. Canonical RNA circularization using group I introns has been achieved with PIE28,54,55, which completes both transesterification steps (Fig. 1a). In the first step of group I intron splicing, exogenous guanosine (exoG), typically guanosine triphosphate (GTP) initiates an attack on the 5’-exon and 5’-intron junction, resulting in RNA cleavage and the addition of a single G at the 5’ end of the 5’-intron (Fig. 1a). Recognizing the potential to bypass the initial step, we directly attempted to omit it and evaluated the feasibility of completing the second step of splicing in PIE independently, leading to the development of a method called PIET (Permuted Intron-Exon through Trans-splicing). This method involves two separate RNAs functioning in trans (Fig. 1b), with the 5’ half-intron and intermediate generated independently in separate IVT reactions, then mixed for further reaction.
a Diagram of PIE method for RNA circularization. b Schematic of the PIET approach for RNA circularization, showing the two components as intermediate and 5’-intron. c Assessment of 5’-intron, GTP, and Mg2+ requirements for PIET. d PIET method schematic comparing RNA circularization using different ratios of 5’ intron to intermediate against PIE. e Illustration of CIRC method for RNA circularization. f Agarose gel results depicting GTP and Mg2+ requirements for CIRC. An impaired group I intron was generated by substituting the first 20 bases of the wild-type group I intron with random sequences. g Agarose gel results displaying RNase R-treated linear precursor and circRNA. h Agarose gel showing linear precursor and circRNA treated with poly(A) polymerase. i FACS results illustrating EGFP expression levels in transfected cells. Data presented as mean ± S.D. (n = 3), with each point representing a biological replicate. An unpaired, two-sided Student’s t-test was used for comparisons. j Cell images taken 24 hr post-transfection with linear precursor or circRNA. The linear RNA in this figure was produced using an impaired group I intron. Source data are provided as a Source Data file.
Intron splicing was observed under different conditions, identifying the 5’-intron and Mg2+ as essential factors, while GTP was not required, thus indicating the omission of the first splicing (Fig. 1c). To verify circRNA production, reverse transcription-polymerase chain reaction (RT-PCR) demonstrated precise exon ligation (Supplementary Fig. 1a), while an RNase R assay further confirmed circRNA presence (Supplementary Fig. 1b). The product, treated with RNase R and poly(A), retained an unchanged circRNA length (Supplementary Fig. 1c). Additionally, transfection of human embryonic kidney (HEK) 293T cells with circRNA harboring an IRES-enhanced green fluorescent protein (EGFP) reporter led to robust expression of EGFP proteins (Supplementary Fig. 1d) and demonstrated higher protein expression levels compared to its linear RNA precursors, indicating sufficient RNA circularization (Supplementary Fig. 1e). These findings collectively confirm that PIET effectively generates circRNA.
PIET operates as a two-component circularization system, allowing for easy manipulation of the 5’-intron-to-intermediate ratio to enhance circularization efficiency. As the ratio was adjusted from 0 to 125, the efficiency improved with an increased supply of the 5’-intron at ratios >5 (Fig. 1d). Although PIET did not surpass PIE in circularization efficiency, its two-component system offers precise control over reaction initiation by enabling the timed addition of the 5’-intron as needed.
Utilizing a complete group I intron is a viable approach for circRNA generation
Given that PIET successfully circularized RNA with a split intron, we hypothesized that a full, unsplit intron could achieve similar circularization (Fig. 1e). This led us to investigate whether an unsplit intron could facilitate efficient RNA circularization by utilizing the second step of splicing as PIET. Intron splicing was observed under various conditions, with Mg2+ identified as a limiting factor, while GTP was not required, indicating that this method bypasses the first step of group I intron splicing (Fig. 1f). RT-PCR confirmed precise exon ligation (Supplementary Fig. 1a), and an RNase R assay validated circRNA production (Fig. 1g). After RNase R treatment, the product was subjected to Poly(A) addition, and circRNA length remained unchanged (Fig. 1h). Similarly, HEK293T cells transfected with circRNA harboring an IRES-EGFP reporter showed efficient protein translation and higher expression levels compared to linear RNA precursors (Fig. 1i, j). These findings collectively confirm the efficacy of this RNA circularization approach.
Additionally, we speculated that the homology arm required in PIE for split intron binding might be unnecessary (Supplementary Fig. 2a). Removing this homology arm significantly enhanced circularization efficiency (Supplementary Fig. 2b). This approach was named CIRC (Complete self-splicing Intron for RNA Circularization) due to its use of an intact intron. Importantly, CIRC differs from the trans-splicing-based circularization method that uses the trans-splicing activity of group I introns49,50,51, as that approach still depends on the first splicing step (Supplementary Fig. 2c).
The number of Gs at the 5’ end influences RNA yield but not circularization efficiency
Before optimizing the construct sequence for CIRC, a key consideration was that the initiation base in CIRC precursors starts from a G at the 5’ end (Fig. 1e). For group I introns whose first base is not G, only one G is typically present at the 5’ end. However, T7 polymerase requires at least 2 Gs at the 5’ end for efficient transcription66,67. Since the Ana intron begins with an A, we investigated the effect of adding extra G at the 5’ end on RNA yield and circularization. To determine whether the additional G affected transcription and circularization, we compared RNAs with 1 G or 2 G extensions at the 5’ end, evaluating RNA yield and circularization efficiency (Supplementary Fig. 2d). The 2 G sample produced a higher RNA yield than the 1 G sample (Supplementary Fig. 2e). While the 1 G sample showed slightly better circularization efficiency than the 2 G sample, the difference was not statistically significant (Supplementary Fig. 2f). To balance RNA yield and circularization efficiency, we used a minimum of 2 Gs at the 5’ end for all CIRC precursors with group I introns in this study.
Sequence optimization for enhanced CIRC efficiency and balanced protein expression
To improve CIRC’s performance, we conducted sequence optimization, focusing on key elements such as exons, homology arm, and spacer sequences—all crucial for effective circularization and tested independently (Fig. 2a). Circularization rate was also critical to assess, as faster rates help preserve RNA integrity. We evaluated circularization at two temperatures, 37°C and 55°C, mainly reflecting circularization rate and maximum circularization efficiency, respectively.
a Diagram of the optimization of CIRC construct, highlighting features examined in subsequent experiments. b−e Gel quantification results showing the effects of exon length (b−d) and homology arm length (e) on circularization efficiency. f FACS results illustrating EGFP expression levels in transfected cells with circRNA constructs containing varying homology arm lengths. g Gel quantification results illustrating the impact of spacer sequence on circularization efficiency. h Schematic of the final optimized CIRC construct. Data are shown as mean ± S.D. (n = 2 or 3), with each point representing a biological replicate. Red denotes circularization at 55°C; blue denotes circularization at 37°C. Source data are provided as a Source Data file.
We then investigated the minimum exon length required for effective circularization, testing 5’- and 3’-exon lengths ranging from 0-nt to 60-nt. When both exons were of equal length, at least 16-nt was required to achieve rapid and efficient circularization (Fig. 2b). Subsequently, we varied the length of one exon while keeping the other constant and found that a total exon length of 27-nt (16-nt for one exon and 11-nt for the other) was sufficient for effective circularization (Fig. 2c, d). For simplicity, we standardized each exon length to 16-nt in further experiments.
We then explored the impact of the homology arm length on circularization, expecting that extended base-pairing would help align splicing sites. As anticipated, a longer homology arm significantly increased both circularization rate and efficiency, reaching nearly 100% efficiency with a homology arm longer than 150-nt (Fig. 2e). However, these sequence alterations are retained in the final circRNA product, potentially affecting protein expression. Testing circRNAs with various homology arm lengths in HEK293T cells, we found that arms longer than 75-nt impaired translation efficiency (Fig. 2f). Adding a spacer before the IRES partially alleviated this, though it didn’t fully restore protein expression (Supplementary Fig. 3a, b). Therefore, we limited the homology arm length to 19-nt to 50-nt to balance assistance with minimal impact on translation.
To mitigate potential conflicts between surrounding sequences and exons during circularization, we tested different spacer sequences, positioning them as either outer or inner spacers (Fig. 2a). To assess this design, linkers of varying lengths were used as outer or inner spacers to circularize the same payload. We observed that the outer spacer accelerated circularization process and performed slightly better in efficiency than the inner spacer (Fig. 2g). Increasing spacer length further enhanced circularization efficiency (Fig. 2g). However, when examining the impact of spacer location on protein expression, the inner spacer showed superior, likely due to its positioning between the IRES and the homology arm (Supplementary Fig. 3c). The reduction in translation observed with the outer spacer group could be alleviated by adding an additional 30-nt AC30 spacer between the homology arm and IRES (Supplementary Fig. 3d). Considering the preference for a relatively short exogenous sequence, an optimized CIRC configuration was developed: a 16-nt exon on each side, a 19- to 50-nt homology arm, and a 30-nt outer spacer, achieving efficient and relatively fast circularization (Fig. 2h). For protein-coding circRNA, we also recommend including an AC30 spacer before the IRES, consistent with previous findings28. This optimized CIRC configuration (with the AC30 spacer before the IRES) demonstrated enhanced circularization without compromising translation efficiency (Supplementary Fig. 3e, f).
The heterogenicity of the 3’ end of RNA poses a challenge for RNA circularization through CIRC
Upon successfully achieving circularization and optimization via CIRC, we sought to identify additional key factors limiting RNA circularization. It has been observed that T7 polymerase-driven IVT often generates a heterogenous 3’ end68,69,70,71. Considering that the last base in the CIRC precursor aligns with the splicing site, we hypothesized that 3’ end homogeneity would significantly influence circularization. It has been reported that using DNA templates with modifications, such as 2’ OMe (2’ methoxy (OCH₃) group), can partially mitigate this effect72. We compared the circularization efficiency of RNAs generated from linearized plasmid, unmodified PCR products, and modified PCR products (Supplementary Fig. 4a). RNA generated from the modified PCR products exhibited the highest circularization efficiency (Supplementary Fig. 4b). Furthermore, it was reported that mutant T7 polymerase can produce RNA with greater 3’ end homogeneity than wild-type T7 polymerase73, may offering a more convenient way for RNA production in CIRC and enhancing circularization efficiency.
CIRC can leverage various self-splicing introns for RNA circularization
In PIE method, introns require additional engineering to identify intron split sites and incorporate homology arms28,58. Since CIRC can facilitate circularization using intact introns (Fig. 1e and Supplementary Fig. 2a), we hypothesized that other natural self-splicing introns could also be utilized to circularize RNA via CIRC. Using the optimized CIRC framework, we matched the intron and corresponding exon sequences to design the linear precursor (Fig. 3a). Initially, we selected a variety of group I introns from databases and previous studies, highlighting their significant diversity65,74,75,76. Potential circRNA generation and efficient intron splicing were observed in most of the selected group I introns, with the exception of those from Closterium tumidum (Ctu) and Trichocoma paradoxa (Tpa) (Fig. 3b).
a Diagram of the CIRC construct incorporating either group I or group II introns. b Agarose gel electrophoresis showing group I intron splicing and the generation of putative circRNA. c Agarose gel electrophoresis showing group II intron splicing and the generation of putative circRNA. d Agarose gel electrophoresis of putative circRNA generated by different group I introns following treatment with poly(A) polymerase. e Left: Quantification of gel data showing circularization efficiency for various introns. Right: FACS results indicating EGFP expression levels. Data are presented as mean ± S.D. (n = 3), with each point representing a biological replicate. Source data are provided as a Source Data file.
Additionally, group II introns without a protein-coding open reading frame (ORF) in the DIV domain (Domain 4) were reported to function independently of proteins, also bypassing the generation of lariat intron77, suggesting their potential for CIRC-mediated RNA circularization. To better simulate the second step of group II intron splicing, we used a monophosphate at the 5’ end of the precursor. Since these group II introns reportedly have in vitro DNA/RNA cleavage activity77, we observed imperfect base-paring in the natural exon binding site (EBS) and intron biding site (IBS) sequences, which could potentially reduce cleavage of the final circRNA. We identified three group II introns from literature and database searches77,78, all of which could achieve putative circRNA generation through CIRC (Fig. 3c). We confirmed the presence of circRNA via poly(A) assays, observing no size change in the circRNA band (Fig. 3d). Upon transfecting these circRNAs into cells, we found that circRNAs led to higher protein expression than the linear RNA from Ctu and Tpa (Fig. 3e). Quantification of RNA circularization was also shown (Fig. 3e). Group II introns did not exhibit cleavage activity under our conditions, maintaining a circRNA band in the poly(A) assay and showing efficient protein expression (Fig. 3d, e).
In summary, we successfully generated protein-coding circRNA using group I and II introns from various origins (Supplementary Table 1). These findings expand the range of introns applicable to CIRC, creating opportunities to identify introns with high-activity or some unique features. Among tested introns, those from Tetrahymena thermophila (Tet) and Azoarcus olearius (Azo) showed high circularization efficiency but did not significantly surpass the performance of the Ana intron. Given the Ana intron’s high efficiency and compatibility with the PIE method, we explored potential advantages of the CIRC method over PIE method, both using the Ana intron. The optimized CIRC with a 19-nt homology arm was compared to the reported PIE construct, which includes a 16-nt 5’ exon, a 52-nt 3’ exon, a 19-nt homology arm, and a ~20-nt linker serving as internal spacers28.
CIRC demonstrates superior circularization efficiency compared to PIE
We began by comparing the circularization efficiency of CIRC and PIE using four different payloads: IRES-EGFP-3′ UTR, IRES-gaussia luciferase (Gluc)-3′ UTR, IRES-firefly luciferase (Fluc)-3’UTR, and POLR2A, with payload sizes ranging from 336 to 2543 nucleotides. In all cases, CIRC demonstrated superior circRNA production compared to PIE (Fig. 4a and Supplementary Fig. 5a).
a Quantified circularization efficiency of RNA in CIRC and PIE from agarose gel electrophoresis data. b Comparison of the recovery rate for the final purified circRNA of the correct size. c, d Comparison of Mg2+ (c) and pH (d) requirements. e, f Comparison of circularization rates at 55°C (e) and during IVT reactions at 37°C (f). Data are shown as mean ± S.D. (n = 3), with each symbol representing a biological replicate. Source data are provided as a Source Data file.
Another critical metric is the final yield of pure circRNA. A common approach to thoroughly eliminate linear RNA is to apply RNase R digestion following poly(A) polymerase treatment39,79,80. Using equal input mass for both CIRC and PIE products, we conducted poly(A) treatment and RNase R digestion, then quantified the final yield of pure circRNA. Analysis of the treated product revealed that CIRC-generated circRNA contained a higher proportion of monomeric circRNAs, appearing as distinct bands corresponding to their expected sizes, whereas PIE displayed a greater proportion of concatemers (Supplementary Fig. 5b).
When comparing the recovery rates of monomeric circRNA, CIRC produced a higher yield than PIE, particularly with EGFP and Gluc payloads, and comparably with Fluc (Fig. 4b). Overall, CIRC showed a modest improvement over PIE, though PIE is highly efficient, making significant enhancements challenging. Consequently, we explored additional advantages of CIRC over PIE.
CIRC performs better than PIE under mild reaction conditions
Effective circularization via self-splicing introns relied on ribozyme activity, requiring optimal Mg2+ concentration, temperature, and reaction time. However, these conditions can promote non-enzymatic RNA degradation, especially in high Mg2+ concentrations, elevated temperatures, and extended incubation times81,82. Additionally, reaction pH affects RNA stability, while higher pH levels increasing RNA hydrolysis risk81. To optimize RNA integrity and circularization efficiency, these factors were systematically tested.
CIRC demonstrated a lower Mg2+ requirement, achieving circularization with only 0.2 mM MgCl2, whereas PIE required around 3.2 mM MgCl2 (Fig. 4c). Furthermore, CIRC demonstrated the ability to perform circularization at a lower pH (Fig. 4d). These findings suggest that CIRC can efficiently circularize RNA while preserving its integrity by operating under lower Mg2+ concentrations and reduced pH conditions. Circularization rates were further evaluated at various Mg2+ concentrations at 55°C with 50 mM 2-(N-morpholino) ethanesulphonic acid (MES) at pH 6.5. At 4 mM Mg2+, CIRC exhibited a faster circularization rate than PIE (Fig. 4e). At 10 mM Mg2+, no significant difference was observed between CIRC and PIE (Supplementary Fig. 5c), while at 25 mM Mg2+, slight inhibition occurred in the second transesterification step of group I intron splicing, leading to faster circularization, lower final circularization efficiency, and the formation of more complex products in PIE (Supplementary Fig. 5d). Additionally, fewer concatemer were formed when using CIRC, indicating reduced intramolecular reactions under these experimental settings (Fig. 4e, Supplementary Fig. 5c, d). RNA circularization efficiency was assessed during IVT reactions at 37°C over various time points using a commercial IVT kit. CIRC achieved faster circularization than PIE in this IVT condition: CIRC reached higher circularization efficiency after 1 hr of IVT than PIE did after 16 hr (Fig. 4f). Although high Mg2+ concentrations, elevated temperature, and prolonged incubation times can compromise RNA integrity83,84,85, this issue appears to be minor for payloads with normal size.
Overall, these unique features of CIRC support better circRNA integrity and improved performance for RNA circularization, which may be particularly important for large RNAs that demand both high circularization efficiency and preserved RNA integrity.
CIRC surpasses the size limitations of circRNA platforms
With its high circularization efficiency and favorable performance under mild conditions, we hypothesized that CIRC could expand the maximum payload size for circRNA. To test this, we conducted IVT using CIRC with a circRNA encoding full-length human dystrophin protein. Initially, an abnormal band of incorrect size was observed, indicating that the correctly sized RNA was compromised (Supplementary Fig. 6a, second lane). We hypothesized that this issue was caused by the high activity of the intact intron, potentially leading to splicing or trans-splicing during transcription. To address this, we aimed to inhibit intron activity during IVT. We tested cyclic guanosine monophosphate (cGMP), a GTP analog lacking a free 3’-OH group, which has been reported as a group I intron splicing inhibitor86. By adding cGMP to reach a final concentration exceeding 22.5 mM, we successfully generated the precursor RNA in CIRC (Supplementary Fig. 6a). Additionally, we formulated a custom IVT buffer that effectively suppressed group I intron activity for large payloads while optimizing Mg2+ concentrations to balance inhibition and IVT RNA yield. For this payload, all tested Mg2+ concentrations successfully inhibited intron activity. However, the 20 mM concentration resulted in low RNA yield, while the 40 mM concentration led to partial RNA degradation (Supplementary Fig. 6b).
With intron activity successfully controlled during IVT, we evaluated CIRC across a range of payload sizes encoding proteins, spanning from 1448 to 12,206 nucleotides (Fig. 5a). For reference, we also presented the sizes of large payloads from other studies, such as SpCas9, truncated DMD, and Factor VIII-EGFP. (Fig. 5a). Spliced introns were observed in all payloads without noticeable impact on RNA integrity (Fig. 5b). For large payloads, distinguishing circRNA from its precursor in gel images proved challenging. To address this, we used RT-qPCR to qualitatively detect circRNA by measuring the abundance of circRNA junctions relative to linear precursors, as direct comparisons of relative abundance across different payloads were not feasible. All payloads showed a significant increase in circRNA junction abundance, indicating successful circRNA generation (Fig. 5c).
a Diagram illustrating payload types and sizes, with cited payloads from previous studies included. b Agarose gel electrophoresis showing spliced intron and RNA integrity of putative circRNA with varying payloads. c Qualitative validation of circRNA using RT-qPCR, since comparisons of the relative abundance of circRNA junctions across different payloads were not feasible. d−h Detection of protein expression for respective payloads: Gluc (d) and Fluc (f) detected with a microplate reader, EGFP (e) detected by FACS, and ABE8e (g) and dystrophin (h) detected by Western blot. Data are shown as mean ± S.D. (n = 3), with each symbol representing a biological replicate. The linear RNA shown in this figure was produced using a modified group I intron with reduced activity. Source data are provided as a Source Data file.
After successfully achieving RNA circularization, we evaluated whether these circRNAs could effectively express proteins. Cells transfected with circRNAs encoding various payloads exhibited robust, full-length protein expression, surpassing that of linear precursor RNAs (Fig. 5d–h). Notably, circRNA encoding human dystrophin protein achieved successful full-length protein expression (Fig. 5h). However, previous reports did not include any protein expression data on their largest construct39,51.
An additional 16,835-nt payload was also tested (Supplementary Fig. 7a), demonstrating intron splicing with high RNA integrity and enriched relative abundance of circRNA junctions (Supplementary Fig. 7b, c). However, full-length protein expression from this construct could not be detected via Western blot, possibly due to low expression levels of the chimera construct.
In summary, CIRC expands the possibilities for circRNA-based applications by overcoming payload size constraints, and the exceptional integrity of large RNAs indicates that CIRC’s capacity has yet to reach its upper limits.
CIRC generates circRNA with minimal extraneous sequences and high efficiency
Immunogenicity is a critical concern for circRNA, with impurities and scar sequences identified as major contributors to this issue29,64. CIRC aims to provide a method for producing high-purity circRNA without scar-related constraints, laying a foundation for investigating the sources of immunogenicity. Scarless circRNA production also aligns with the objective of replicating endogenous circRNA sequences. Prior to evaluating CIRC’s ability to generate scarless circRNA, we examined the scar elements in the optimized CIRC construct, which include exon, homology arm, and spacer sequences for efficient circularization. To minimize extraneous sequences, we removed both the homology arm and spacer sequence and shortened the exon sequence, aiming to utilize sequences commonly present in any RNA to facilitate easy swapping for circularization (Fig. 6a).
a Diagram illustrating the principle of generating scarless circRNA using CIRC. b Minimal exon sequence requirements when using the Ana intron in CIRC. c Diagram showing the programmable P1 stem within the Ana intron in CIRC. d Quantified circularization efficiency from agarose gel electrophoresis data for constructs shown in (c). e Diagram illustrating the approach to generate scarless circRNA by using an internal structure as the homology arm. f Quantified circularization efficiency from agarose gel electrophoresis data for scarless circRNA generation using various split sites within CVB3 IRES in CIRC. g A diagram illustrating the workflow for the detection of circRNA immunogenicity, created in BioRender. Shen, Y. (2025) https://BioRender.com/j5i3pnq. h–j qPCR results showing the relative expression of RIG-I, IL-6, and TNF-α following RNA transfection into A549 cells, serving as indicators of immunogenicity. Data are shown as mean ± S.D. (n = 3), with each symbol representing a biological replicate. Source data are provided as a Source Data file.
The Ana intron has demonstrated functionality with short exon sequences87, so we tested CIRC using minimal exons and found that a 3-nt 5’ exon (CUU) and a 2-nt 3’ exon (AA) supported circularization with partial loss of splicing activity (Fig. 6b). This configuration revealed that a “CUUAA” motif within the payload can be used for scarless circRNA generation. Additionally, the 5’-exon sequence base-pairs with the intron to form the P1 structure of group I intron, allowing for sequence adjustments while preserving base-pairing and intron activity. In the 3-nt 5’-exon sequence, the final base of the 5’-exon forms a G-U wobble pair, which is considered crucial for the splicing activity of group I introns88. We tested all 16 alternative constructs by varying the other two bases in the 5’ exon, using a short payload POLR2A to assess their impact on splicing (Fig. 6c). The results indicated that all 16 pairs were capable of intron splicing with varying splicing efficiency (Fig. 6d). Thus, locating a “NNUAA” motif (or “NNUA” for potential reduced efficiency) in any RNA payload enables scarless circRNA generation. However, the effectiveness of other NNU motifs for RNA circularization should be carefully evaluated with specific sequence contexts.
Beyond exon sequences, the removal of the homology arm and spacer might reduce circularization efficiency. Therefore, alternative strategies need to be explored to enhance circularization efficiency. Since these sequences aid in splicing site accessibility, we considered that secondary structures within the payload itself might serve a similar role (Fig. 6e). Given the highly structured nature of IRES, we applied the CIRC method while identifying splitting sites within the CVB3 IRES sequence. Sixteen different “NNUA” motif split sites within the IRES were evaluated, with three demonstrating exceptionally high splicing efficiency, highlighting the versatility of this strategy in generating scarless circRNAs (Fig. 6f). Among them, the split site at position 403, utilizing a UCU/AA motif, is recommended for generating scarless circRNAs containing the CVB3 IRES. By defining the minimal exon requirements and eliminating extra homology arms and spacers, the scar length was significantly reduced. The remaining scar sequence (NNUAA or NNUA in this case) could be located and replaced within the payload sequence to produce scarless circRNA.
CIRC produces circRNA with minimal immunogenicity
After achieving scarless circRNA production, we assessed the immunogenicity of circRNA with different scar lengths produced by CIRC in cells (Fig. 6g). Observation from previous studies indicated that purity, scar length, transfection dose, and phosphatase treatment influence immunogenicity. Initially, we synthesized the reported IRES-mCherry circRNA using the optimized CIRC construct64 and evaluated its immunogenicity at various doses, with and without phosphatase treatment, using modified or unmodified linear RNA and poly(I:C) as controls. Post-phosphatase treatment, circRNA exhibited low immunogenicity (Supplementary Fig. 8), showing similar results as previous report29. Besides, with doses over 0.1 μg per 24-well plate was sufficient to reveal immunogenic differences (Supplementary Fig. 8). We further explored the immunogenicity of phosphatase-treated circRNA with different scar lengths and added T4 RNA ligase-generated circRNA as a control, known for low immunogenicity64. However, no direct correlation was observed between scar length and immunogenicity, as all circRNAs exhibited low immunogenicity compared to unmodified linear RNA or poly(I:C) (Fig. 6h), suggesting that scar length has a minimal impact on circRNA immunogenicity. However, structure distortions caused by scars in specific sequence contexts may significantly influence immunogenicity89 and should be carefully evaluated on a case-by-case basis.
Additional phosphatase-treated circRNAs generated using CIRC were also evaluated. Circular RNA circPOLR2A, known for its low immunogenicity and therapeutic potential as a PKR inhibitor39,64,90, was successfully produced as a scarless circRNA using CIRC. This scarless circPOLR2A exhibited minimal immunogenicity, comparable to circPOLR2A generated by T4 RNA ligase (Fig. 6i). Furthermore, three additional payloads circularized with the optimized CIRC construct were tested for immunogenicity, and all demonstrated consistently low immunogenicity (Fig. 6j). These findings underscore CIRC as an effective platform for generating circRNA with minimal scar constraints and low immunogenicity.
CIRC simplifies circRNA purification with RNase R
Unlike linear mRNA production, self-splicing intron-based circRNA generation produces additional byproducts, such as linear precursors and spliced introns, necessitating efficient circRNA purification methods for therapeutic applications. RNase R, a 3’ exoribonuclease, is commonly used in laboratories due to its effectiveness and simplicity in circRNA purification91,92. Comparing CIRC to PIE, we hypothesized that differences in RNase R resistance among linear precursors arise from variations in their 3’ ends. In PIE, the 3’ end of the linear precursor is embedded within the homology arm, a feature critical for RNA circularization28 but one that may increase resistance to RNase R digestion (Fig. 7a). Conversely, the 3’ end of CIRC’s precursor consists of the simpler 5’-exon structure, potentially making it more susceptible to RNase R digestion (Fig. 7a).
a Diagram depicting RNase R sensitivity of precursors in CIRC and PIE. b Agarose gel electrophoresis showing RNase R treatment of precursors in CIRC and PIE over different time periods. c Agarose-gel electrophoresis showing RNase R treatment of the 2-hr and 4-hr IVT RNA product in CIRC over varying durations. d Top: Diagram illustrating poly(A) insertion within the intron in CIRC. Bottom: Workflow for oligo(dT) column-mediated circRNA purification. e Agarose gel electrophoresis showing the effect of different number of As insertion on circularization (f) Agarose gel electrophoresis of the flow-through from the oligo(dT) column. In (b−f) no additional replicate was done. Source data are provided as a Source Data file.
When we treated the precursors from both methods with RNase R over varying durations, the PIE precursor showed greater resistance (Fig. 7b and Supplementary Fig. 9a, b). Additionally, it has been documented that the Ana intron exhibits self-ligase activity, allowing the intron ends to ligate93. Our observations showed that the 5’ and 3’ ends of the intron ligated during prolonged IVT reactions or RNA circularization, resulting in the formation of a circular intron in CIRC and a bound intron in PIE, both of which resisted RNase R digestion (Supplementary Fig. 9c). Preventing intron ligation while achieving RNA circularization thus emerged as an effective strategy to enhance RNase R efficiency. Notably, CIRC allows for rapid circularization without intron ligation during IVT (Fig. 4f). To enhance RNase R digestion efficiency, we selected RNA samples shortly after initiating IVT using CIRC (Fig. 4f, IVT for 2 hr and 4 hr), which demonstrated effective intron digestion (Fig. 7c). In summary, CIRC’s expedited circularization process minimizes intron ligation through shortened reaction time, allowing for more effective RNase R digestion. Overall, CIRC demonstrated greater suitability for streamlined RNA purification with RNase R, offering a convenient approach for obtaining highly pure circRNA.
CIRC enables oligo(dT)-based circRNA purification
Canonical circRNA purification methods depend on RNA exonucleases like RNase R79, size-based separation techniques like size exclusion chromatography (SEC)29, or specialized materials like wood-derived macroporous cellulose (WMC)94. In contrast, mRNA purification commonly employs oligo(dT) columns, which selectively bind to poly(A) tails95,96,97, offering a size-independent purification strategy. Considering that CIRC’s byproducts contain introns, we tested whether inserting poly(A) sequences into the Ana group I intron’s P6 domain would allow oligo(dT)-based removal of byproducts (Fig. 7d). To validate, we used RNA mixes with low circularization efficiency—achieved by employing short exons lacking spacer sequences—and evaluated the impact of poly(A) insertions up to 75-nt on splicing efficiency. The results showed only minimal impairment of splicing (Fig. 7e). Subsequently, we incubated a mix of circRNA and byproducts with oligo(dT) magnetic beads and collected the supernatant to assess circRNA enrichment. When the A insertions exceeded 25-nt, significant circRNA enrichment was observed in the supernatant (Supplementary Fig. 9d). Since the precursor was harder to remove due to its larger size, multiple rounds of bead incubation were effective, with three rounds completely removing both precursor and intron when the A insertions reached 34- and 75-nt (Supplementary Fig. 9e).
Following this proof of concept, we applied a commercial oligo(dT) column to increase RNA purification throughput. We loaded RNA, both with and without A insertions in the intron, onto a commercial oligo(dT) column (Fig. 7d). The flow-through was collected and analyzed on an agarose gel. Encouragingly, the RNA was successfully purified, with most impurities removed, enabling efficient single-round purification (Fig. 7f). In summary, CIRC proved compatible with oligo(dT)-based RNA purification, offering a convenient and efficient method for obtaining high-purity circRNA. Although the insertion of a poly(A) sequence resulted in minimal impairment of intron activity, the ability to obtain high-purity circRNA with ease made this trade-off acceptable.
Discussion
This study presents two in vitro RNA circularization methods: PIET and CIRC, which utilize the second step of self-splicing intron processing to provide efficient methods for RNA circularization. CIRC has been optimized for high efficiency and demonstrates several advantages over the commonly used PIE, particularly in circularization efficiency and RNA integrity preservation (Figs. 2 and 4). Notably, CIRC is not limited by intron splitting and can leverage various natural group I and group II introns for RNA circularization, enabling the discovery of potentially superior introns for this process (Fig. 3). Additionally, exploring the diversity of self-splicing introns may further enable RNA circularization under diverse conditions, including in vivo.
Current limitations in generating circRNA in vitro with payloads over 9000-nt28,39,51 restrict the broader application of the circRNA platform for large protein expression or loading multiple payloads within a single circRNA. Through advancements in CIRC, we extend the payload size limit, achieving circularization of large RNA while preserving RNA integrity (Fig. 5). This progress could expand the applications of circRNA, such as developing new protein replacement therapies for diseases caused by loss of function in large, full-length proteins, including DMD and Usher syndrome.
Furthermore, ribozymes-mediated RNA circularization often leaves undesired scar sequences in circRNA, which are considered immunogenic and complicate the production of endogenous circRNA with identical sequences25,64. In contrast, CIRC enables efficient RNA circularization with minimal constraints on scar length (Fig. 6). We examined the impact of scars on immunogenicity and found no clear correlation, although further studies are needed to assess how structural distortions from scars may influence immunogenicity89.
An important area for future optimization of the circRNA platform involves introducing chemical modifications as well as enabling protein expression within circRNA44,45. However, for protein-coding circRNA, modified IRES elements often lose their ability to initiate translation29. Identifying modification-tolerant IRES, directly incorporating specific caps or modifications into circRNA, and exploring alternative mechanisms for translation initiation are necessary to address this challenge. Recent studies have made progress in tackling this issue by incorporating cap structure within modified circRNA44,45. However, limitations remain on convenience and efficiency in the circRNA generation. Therefore, efficient circularization methods compatible with modified bases are needed. For ribozymes-based methods, certain RNA modifications can hinder ribozyme activity, limiting the ability to circularize modified RNA bases29,48. While trans circularization method utilizing unmodified ribozyme has been also developed, they do not resolve the issue of protein translation in modified circRNA51. To fully unlock the potential of circRNA-based therapeutics, it is essential to coordinate solutions for translation within modified circRNAs while developing scalable and efficient circularization methods. Advancements in these areas would significantly enhance the versatility of circRNA platform, paving the way for its widespread applications.
Methods
Plasmids construction
The sequences involved PCR amplification of sequences from our lab’s plasmids or sequences synthesized by Tsingke Biotech. Cloning was performed using Gibson assembly or enzyme digestion and ligation. All constructs were cloned into the pUC57-Kan backbone between the M13 forward and M13 reverse primer sites, downstream of a T7 promoter. A guide to using the CIRC-V1 construct is provided in the Supplementary Note. Detailed sequence information is provided in the Supplementary Data 1. Gibson assembly was conducted with Gibson Assembly Master Mix (New England Biolabs (NEB), E2611L) or 2× MultiF Seamless Assembly Mix (Abclonal, RK21020) according to the manufacturer’s instructions. Restriction enzymes from NEB and the T4 DNA ligase (NEB, M0202L) were used for digestion and ligation. Trans1-T1 Phage Resistant Chemically Competent Cell (Transgene, CD501-03) was used for transformation.
DNA template generation
Linearized plasmids were produced using endonuclease such as BbsI-HF (NEB, R3539L), BsmBI-v2 (NEB, R0739L) or BsaI-HFv2 (NEB, R3733L) for CIRC and PmeI (NEB, R0560L) or AscI (NEB, R0558L) for PIE. Linearized plasmids were purified by DNA Clean & Concentrator (Zymo, D4014). Primers with 2’ OMe modification were obtained from Tsingke Biotech. PCR products were amplified using specific primers with PrimeSTAR GXL Premix (Takara, R051A) and purified by gel recovery using Zymoclean Gel DNA Recovery Kit (ZYMO, D4008).
In vitro transcription
In vitro transcriptions were conducted using the HiScribe™ T7 High Yield RNA Synthesis Kit (NEB, E2040S), using a final concentration of NTPs to be 7.5 mM each. The cGMP used for inhibition of intron splicing was added to the IVT reaction initially. Homemade 10× IVT buffer contained 400 mM HEPES (pH = 6.8), 200−400 mM MgCl2, 20 mM DL-Dithiothreitol (DTT), and 10 mM spermidine, and was used for short-time IVT reactions to prevent intron splicing.
Purification of circRNA
Purification of circRNA involved treating IVT products with DNase I (NEB, M0303L), followed by column purification using the Monarch RNA Cleanup Kit (NEB, T2040L). For samples conducting additional circularization, circRNAs were finally column purified and concentrated with the RNA Clean & Concentrator Kit (ZYMO, R1018) for agarose gel electrophoresis analysis or transfection.
Additional circularization using self-splicing introns
In certain cases, RNA underwent an additional circularization process using different custom-made buffers at different temperatures and incubation times. HEPES buffers with a pH range of 6.8 to 8.0 were procured from HARVERBIO, and MES buffers with a pH range of 5.0 to 6.5 were obtained from PERFEMIKER. For clarity, the circularization conditions applied in each figure are detailed in Table 1, with corresponding Supplementary Figures. using the same reaction conditions. For RNA circularization using PIE, GTP was added to a final concentration of 2 mM.
RNA agarose gel electrophoresis
RNA agarose gel electrophoresis was performed using Tris-Acetate-EDTA (TAE) buffer with in-house-prepared agarose gels. Agarose concentrations ranged from 1% to 2%, and 0.5× or 1× TAE buffer was used for RNA electrophoresis. Gel data were analyzed using Image Lab v6.1. Uncropped and unprocessed images are available in Source Data or Supplementary Information.
Calculation of circularization efficiency and recovery rate
RNA agarose gel electrophoresis images were analyzed using Image J v1.53 k to measure the integrated density of bands in each lane. Circularization efficiency was calculated using the formula: ((Integrated density of circRNA)/(length of circRNA)) / (((Integrated density of precursor)/(length of precursor)) + ((Integrated density of circRNA) / (length of circRNA))). To calculate the recovery rate, equal amounts (μg) of input RNA mixtures were treated with poly(A) polymerase following RNase R digestion under the same conditions. The recovery rate was calculated using the formula: (amount of undigested circRNA (μg)) / (input RNA (μg)). Data analysis was performed using Graphpad Prism v10.1.2.
Cell culture
HEK293T (ATCC, CRL-3216) and A549 (CCL-185) were maintained in our lab, with Dulbecco’s modified eagle medium (DMEM) (Gibco, C11965500BT), supplemented with 10% fetal bovine serum (FBS) (Biological Industrials, C04001-500), 1% penicillin-streptomycin (with a final concentration of 5 mg/mL and 10 mg/mL respectively), 1% GlutaMAX (Gibco, 35050061), and 1% NEAA (Non-Essential Amino Acids Solution) (Gibco, 11140050) in a 5% CO2 incubator at 37°C.
RNA transfection
For RNA transfection, HEK293T cells were seeded at ~1.5 × 105 cells per well in 24-well plates, 3 × 105 cells per well in 12-well plate. After 24 hr, RNA was transfected into the cells using Lipofectamine MessengerMax (Invitrogen, LMRNA008). 24 or 60 hr post transfection, the cells were collected for the subsequent detections. A549 cells were seeded at ~8 × 104 cells per well in 24-well plates. After 24 hr, RNA or Poly(I:C) (Sigma, P9582) was transfected into the cells using Lipofectamine Messenger Max. 6 hr post transfection, the cells were harvested for subsequent analyses.
RNase R treatment
RNase R treatment involved heating RNA at 65°C for 5 min, followed by cooling on ice. Reaction buffer and RNase R (mostly 0.2 U/μg) were added and mixed (Epicentre, RNR07250), and the reaction was conducted at 37°C. RNA was then purified and concentrated with the RNA Clean & Concentrator Kit (ZYMO, R1018).
Poly(A) tailing of RNA
RNA was heated at 65°C for 5 min, followed by cooling on ice. Reaction buffer, ATP, polymerase, and appropriate RNAs were mixed and reacted according to the manufacturer’s instruction (NEB, M0276L).
RT-PCR detection of the circRNA junctions
CircRNAs were reverse transcribed into cDNA using the AMV First Strand cDNA Synthesis Kit (Sangon, B532445). Target sequences were amplified via PCR using specific primers, and the PCR products were cloned into T vectors using the Zero TOPO-TA/Blunt Cloning Kit (ABclonal, RK30130). Single clones were selected for Sanger sequencing. For quantifying circRNA junction abundance (Fig. 5), reverse transcription was performed using the SuperScript IV First-Strand Synthesis System (Thermo, 18091050), followed by qPCR detection using TB Green Premix Ex Taq (Tli RNase H Plus) (Takara, RR42LR) on a CFX Connect Real-Time PCR Detection System (Bio-Rad). Sequences of qPCR primers were included in Supplementary Information. Data was analyzed using Graphpad Prism v10.1.2.
Fluorescence activated cell sorting (FACS) analysis
Cells were trypsinized and resuspended in phosphate buffered saline (PBS) containing 2% FBS. Samples were analyzed using an LSRFortessa cytometer (BD Biosciences), and data were processed with FlowJo software. The gating strategy was shown in Supplementary Fig. 10. At least 2 × 104 singlets cells were collected for analysis. Data were analyzed using Flowjo v10.8.1.
Selection of group I and group II introns
Group I and group II introns were selected from the reported database74,78 based on specific sequence information. Introns were ranked by Minimal Free Energy (MFE) and normalized to sequence length. Top-ranked sequences and group II introns from previous studies77 were tested for in vitro activity. Active group I introns were subsequently applied to CIRC. The origins of the introns used are listed in Supplementary Information as Supplementary Table 1.
Luciferase activity detection
Gaussia and firefly luciferase activities were measured using the Gaussia Luciferase Reporter Gene Assay Kit (Beyotime, RG021M) or britelite plus (PerkinElmer, 6066769) following the manufacturer’s protocols. Data were collected using an Infinite M200 (TECAN) or Spectra i3 max (Molecular Device). Data were analyzed using Graphpad Prism v10.1.2.
Western blot
After transfection, cells were lysed with cell lysis buffer (Macklin, C874806) supplemented with 1% protease inhibitor cocktail (MCE, HY-K0010). Cells were trypsinized, centrifuged to form a pellet, resuspended in lysis buffer, and incubated on ice for 30 min. After centrifugation at 12,000 × g for 10 min, the supernatant was collected. Samples were prepared with 5× reducing SDS-PAGE loading buffer (CwBio, CW0027S) and boiled at 99°C for 10 min. Proteins were separated by SDS-PAGE using SurePAGE™ (Genscript, M00654) and transferred to membranes using the TransBlot® Turbo™ Transfer System (Bio-Rad) under a constant current of 1.0 A for 30 min. Blocking was carried out with QuickBlock™ Blocking Buffer (Beyotime, P0252). Primary antibodies specific to dystrophin (Abclonal, A1411, 1:250 dilution), vinculin (Abclonal, A2752, ARC51900, 1:10,000 dilution), tubulin (Cwbio, CW0098M, 3G7, 1:5000 dilution), Cas9 (Beyotime, AF0123, 1:5000 dilution), and Flag (Easybio, BE2004, 1:10,000 dilution) were diluted in QuickBlock™ Primary Antibody Dilution Buffer for Western Blot (Beyotime, P0256). Secondary antibodies from Jackson Immune Research were diluted in QuickBlock™ Secondary Antibody Dilution Buffer (Beyotime, P0258). Washing steps were performed using home-made PBST (1% Tween-20 in PBS). Detection was conducted using Clarity™ Western ECL substrate (Bio-Rad, 1705060) and imaged with a Bio-Rad ChemiDoc system. Uncropped and unprocessed scans were provided in Source Data.
RNA circularization using T4 RNA ligase
Scarless circPOLR2A was synthesized using T4 RNA Ligase (Thermo, EL0021) according to the manufacturer’s instruction. Scarless circRNA of IRES-mCherry was generated using T4 RNA Ligase 2 (NEB, M0329L), as previously reported23. In brief, by using internal secondary structure within CVB3 IRES without additional DNA splint, RNA nick within dsRNA region was formed as the substrate for T4 RNA Ligase 2.
RNA extraction and RT-qPCR for immune-related genes expression
RNA was extracted using the Quick-RNA Miniprep Kit (Zymo, R1055). Reverse transcription was performed with the ABScript III RT Master Mix for qPCR (Abclonal, RK20428). qPCR was conducted using TB Green Premix Ex Taq (Tli RNase H Plus) (Takara, RR42LR) on a Roche 480 LightCycler. The relative expressions of RIG-I, IL-6, TNF-α are normalized with 18S rRNA. Sequences of qPCR primers were included in Supplementary Information. Data were analyzed using Graphpad Prism v10.1.2.
RNA purification by oligo(dT) magnetic beads
VAHTS mRNA Capture Beads (Vazyme, N401-02) was used for circRNA purification by depleting poly(A) containing byproducts. In brief, 2 μg RNA was incubated with 50 μL beads for one round, and the supernatant after incubation was used for downstream analysis or another round of incubation.
RNA purification by oligo(dT) column
A NmSCREEN 4.7 mL oligo(dT) column (Nanomicron, 77105-17030-05150) was used in remove poly(A)-containing byproducts for circRNA purification. Samples were loaded at 50°C in a binding buffer containing 50 mM Tris-HCl (pH = 7.5), 500 mM NaCl, and 5 mM EDTA. Elution was performed at 65°C using nuclease-free water.
Statistics and reproducibility
n represents the number of independent experiments performed in parallel. For data represented comparison between groups and calculating p-value, n = 3 was chosen as sample size. Unpaired two-tailed Student’s t-test was implemented for group comparisons as indicated in the figure legends. No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated in this study are included in the Source Data file or the Supplementary Information. Source data are provided with this paper.
References
-
Damase, T. R. et al. The limitless future of RNA therapeutics. Front Bioeng. Biotechnol. 9, 628137 (2021).
-
Zhu, Y., Zhu, L., Wang, X. & Jin, H. RNA-based therapeutics: an overview and prospectus. Cell Death Dis. 13, 644 (2022).
-
Kim, Y. K. RNA therapy: rich history, various applications and unlimited future prospects. Exp. Mol. Med. 54, 455–465 (2022).
-
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
-
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines – a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
-
Chaudhary, N., Weissman, D. & Whitehead, K. A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov. 20, 817–838 (2021).
-
Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020).
-
Baden, L. R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021).
-
Lang, F., Schrors, B., Lower, M., Tureci, O. & Sahin, U. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat. Rev. Drug Discov. 21, 261–282 (2022).
-
Rojas, L. A. et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature 618, 144–150 (2023).
-
Sahin, U., Kariko, K. & Tureci, O. mRNA-based therapeutics-developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014).
-
Koeberl, D. et al. Interim analyses of a first-in-human phase 1/2 mRNA trial for propionic acidaemia. Nature 628, 872–877 (2024).
-
Rothgangl, T. et al. In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat. Biotechnol. 39, 949–957 (2021).
-
Musunuru, K. et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593, 429–434 (2021).
-
Gillmore, J. D. et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021).
-
Krienke, C. et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science 371, 145–153 (2021).
-
Hotz, C. et al. Local delivery of mRNA-encoded cytokines promotes antitumor immunity and tumor eradication across multiple preclinical tumor models. Sci. Transl. Med. 13, eabc7804 (2021).
-
Rurik, J. G. et al. CAR T cells produced in vivo to treat cardiac injury. Science 375, 91–96 (2022).
-
de Picciotto, S. et al. Selective activation and expansion of regulatory T cells using lipid encapsulated mRNA encoding a long-acting IL-2 mutein. Nat. Commun. 13, 3866 (2022).
-
Hunter, T. L. et al. In vivo CAR T cell generation to treat cancer and autoimmune disease. Science 388, 1311–1317 (2025).
-
Rosa, S. S., Prazeres, D. M. F., Azevedo, A. M. & Marques, M. P. C. mRNA vaccines manufacturing: challenges and bottlenecks. Vaccine 39, 2190–2200 (2021).
-
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021).
-
Qu, L. et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 185, 1728–1744.e1716 (2022).
-
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J. & Kleinschmidt, A. K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 73, 3852–3856 (1976).
-
Liu, C. X. & Chen, L. L. Circular RNAs: characterization, cellular roles, and applications. Cell 185, 2016–2034 (2022).
-
Enuka, Y. et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44, 1370–1383 (2016).
-
Zhang, Y. et al. The biogenesis of nascent circular RNAs. Cell Rep. 15, 611–624 (2016).
-
Wesselhoeft, R. A., Kowalski, P. S. & Anderson, D. G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9, 2629 (2018).
-
Wesselhoeft, R. A. et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74, 508–520.e504 (2019).
-
Chen, C. K. et al. Structured elements drive extensive circular RNA translation. Mol. Cell 81, 4300–4318.e4313 (2021).
-
Fan, X., Yang, Y., Chen, C. & Wang, Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat. Commun. 13, 3751 (2022).
-
Chen, R. et al. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 41, 262–272 (2023).
-
Amaya, L. et al. Circular RNA vaccine induces potent T cell responses. Proc. Natl. Acad. Sci. USA 120, e2302191120 (2023).
-
Feng, Z. et al. An in vitro-transcribed circular RNA targets the mitochondrial inner membrane cardiolipin to ablate EIF4G2(+)/PTBP1(+) pan-adenocarcinoma. Nat. Cancer 5, 30−46 (2023).
-
Zhang, Y. et al. Small circular RNAs as vaccines for cancer immunotherapy. Nat. Biomed. Eng. 9, 249–267 (2025).
-
Jing, W. et al. An in situ engineered chimeric IL-2 receptor potentiates the tumoricidal activity of proinflammatory CAR macrophages in renal cell carcinoma. Nat. Cancer 6, 838–853 (2025).
-
Wang, Y. et al. Synergically enhanced anti-tumor immunity of in vivo CAR by circRNA vaccine boosting. Cell Rep. Med. https://doi.org/10.1016/j.xcrm.2025.102250 (2025).
-
Yi, Z. et al. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol. 40, 946–955 (2022).
-
Guo, S. K. et al. Therapeutic application of circular RNA aptamers in a mouse model of psoriasis. Nat. Biotechnol. 43, 236–246 (2024).
-
Feng, X. et al. Circular RNA aptamers targeting neuroinflammation ameliorate Alzheimer disease phenotypes in mouse models. Nat. Biotechnol. https://doi.org/10.1038/s41587-025-02624-w (2025).
-
Guo, S. K. et al. Therapeutic circRNA aptamer alleviates PKR-associated osteoarthritis. Sci. Bull. (Beijing) 70, 2232−2236 (2025).
-
Micura, R. Cyclic oligoribonucleotides (RNA) by solid-phase synthesis. Chem.-Eur. J. 5, 2077–2082 (1999).
-
Muller, S. & Appel, B. In vitro circularization of RNA. RNA Biol. 14, 1018–1027 (2017).
-
Chen, H. et al. Chemical and topological design of multicapped mRNA and capped circular RNA to augment translation. Nat. Biotechnol. 43, 1128–1143 (2024).
-
Fukuchi, K. et al. Internal cap-initiated translation for efficient protein production from circular mRNA. Nat. Biotechnol. https://doi.org/10.1038/s41587-025-02561-8 (2025).
-
Schindewolf, C., Braun, S. & Domdey, H. In vitro generation of a circular exon from a linear pre-mRNA transcript. Nucleic Acids Res. 24, 1260–1266 (1996).
-
Chen, H. et al. Preferential production of RNA rings by T4 RNA ligase 2 without any splint through rational design of precursor strand. Nucleic Acids Res. 48, e54 (2020).
-
Su, C. I. et al. A cis-acting ligase ribozyme generates circular RNA in vitro for ectopic protein functioning. Nat. Commun. 15, 6607 (2024).
-
Cui, J. et al. A precise and efficient circular RNA synthesis system based on a ribozyme derived from Tetrahymena thermophila. Nucleic Acids Res. 51, e78 (2023).
-
Lee, K. H. et al. Efficient circular RNA engineering by end-to-end self-targeting and splicing reaction using Tetrahymena group I intron ribozyme. Mol. Ther. Nucleic Acids 33, 587–598 (2023).
-
Du, Y. et al. Efficient circular RNA synthesis for potent rolling circle translation. Nat. Biomed. Eng. 9, 1062–1074 (2024).
-
Petkovic, S. & Muller, S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 43, 2454–2465 (2015).
-
Obi, P. & Chen, Y. G. The design and synthesis of circular RNAs. Methods 196, 85–103 (2021).
-
Puttaraju, M. & Been, M. D. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 20, 5357–5364 (1992).
-
Ford, E. & Ares, M. Jr. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc. Natl. Acad. Sci. USA 91, 3117–3121 (1994).
-
Li, H. et al. Circular RNA cancer vaccines drive immunity in hard-to-treat malignancies. Theranostics 12, 6422–6436 (2022).
-
Li, H. et al. Intranasal prime-boost RNA vaccination elicits potent T cell response for lung cancer therapy. Signal Transduct. Target Ther. 10, 101 (2025).
-
Liao, K. C. et al. Characterization of group I introns in generating circular RNAs as vaccines. Nucleic Acids Res. 53, gkaf089 (2025).
-
Qi, S. et al. Efficient circularization of protein-encoding RNAs via a novel cis-splicing system. Nucleic Acids Res. 52, 10400–10415 (2024).
-
Chen, C. et al. A flexible, efficient, and scalable platform to produce circular RNAs as new therapeutics. BioRxiv https://doi.org/10.1101/2022.05.31.494115 (2022).
-
Gao, X., Chen, K. & Wang, H. NicOPURE: nickless RNA circularization and one-step purification with engineered group II introns and cyclizing UTRs. Nucleic Acids Res. 53, gkaf310 (2025).
-
Muller, U. F. Design and experimental evolution of trans-splicing group I intron ribozymes. Molecules 22, 75 (2017).
-
Chen, Y. G. et al. Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238.e225 (2017).
-
Liu, C. X. et al. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell 82, 420–434.e426 (2022).
-
Kruger, K. et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31, 147–157 (1982).
-
Komura, R., Aoki, W., Motone, K., Satomura, A. & Ueda, M. High-throughput evaluation of T7 promoter variants using biased randomization and DNA barcoding. PLoS ONE 13, e0196905 (2018).
-
Conrad, T., Plumbom, I., Alcobendas, M., Vidal, R. & Sauer, S. Maximizing transcription of nucleic acids with efficient T7 promoters. Commun. Biol. 3, 439 (2020).
-
Milligan, J. F. & Uhlenbeck, O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180, 51–62 (1989).
-
Triana-Alonso, F. J., Dabrowski, M., Wadzack, J. & Nierhaus, K. H. Self-coded 3’-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J. Biol. Chem. 270, 6298–6307 (1995).
-
Zaher, H. S. & Unrau, P. J. T7 RNA polymerase mediates fast promoter-independent extension of unstable nucleic acid complexes. Biochemistry 43, 7873–7880 (2004).
-
Gholamalipour, Y., Karunanayake Mudiyanselage, A. & Martin, C. T. 3’ end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character-RNA-Seq analyses. Nucleic Acids Res. 46, 9253–9263 (2018).
-
Kao, C., Zheng, M. & Rudisser, S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5, 1268–1272 (1999).
-
Dousis, A., Ravichandran, K., Hobert, E. M., Moore, M. J. & Rabideau, A. E. An engineered T7 RNA polymerase that produces mRNA free of immunostimulatory byproducts. Nat. Biotechnol. 41, 560–568 (2023).
-
Zhou, Y. et al. GISSD: Group I intron sequence and structure database. Nucleic Acids Res. 36, D31–D37 (2008).
-
Cech, T. R., Zaug, A. J. & Grabowski, P. J. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 27, 487–496 (1981).
-
Dolan, G. F. & Muller, U. F. Trans-splicing with the group I intron ribozyme from Azoarcus. RNA 20, 202–213 (2014).
-
Liu, Z. X. et al. Hydrolytic endonucleolytic ribozyme (HYER) is programmable for sequence-specific DNA cleavage. Science 383, eadh4859 (2024).
-
Candales, M. A. et al. Database for bacterial group II introns. Nucleic Acids Res. 40, D187–D190 (2012).
-
Xiao, M. S. & Wilusz, J. E. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3’ ends. Nucleic Acids Res. 47, 8755–8769 (2019).
-
Xiao, M. S. & Wilusz, J. E. Purification of circular RNAs using poly(A) tailing followed by RNase R digestion. Methods Mol. Biol. 2765, 3–19 (2024).
-
Chheda, U. et al. Factors affecting stability of RNA – temperature, length, concentration, pH, and buffering species. J. Pharm. Sci. 113, 377–385 (2024).
-
Fabre, A. L., Colotte, M., Luis, A., Tuffet, S. & Bonnet, J. An efficient method for long-term room temperature storage of RNA. Eur. J. Hum. Genet 22, 379–385 (2014).
-
Li, Y. F. & Breaker, R. R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 121, 5364–5372 (1999).
-
Guth-Metzler, R. et al. Cutting in-line with iron: ribosomal function and non-oxidative RNA cleavage. Nucleic Acids Res. 48, 8663–8674 (2020).
-
Guth-Metzler, R. et al. Goldilocks and RNA: where Mg2+ concentration is just right. Nucleic Acids Res. 51, 3529–3539 (2023).
-
Panchapakesan, S. S. S. & Breaker, R. R. The case of the missing allosteric ribozymes. Nat. Chem. Biol. 17, 375–382 (2021).
-
Zaug, A. J., McEvoy, M. M. & Cech, T. R. Self-splicing of the group I intron from Anabaena pre-tRNA: requirement for base-pairing of the exons in the anticodon stem. Biochemistry 32, 7946–7953 (1993).
-
Doudna, J. A., Cormack, B. P. & Szostak, J. W. RNA structure, not sequence, determines the 5’ splice-site specificity of a group I intron. Proc. Natl. Acad. Sci. USA 86, 7402–7406 (1989).
-
Liu, C. X., Yang, L. & Chen, L. L. Dynamic conformation: marching toward circular RNA function and application. Mol. Cell 84, 3596–3609 (2024).
-
Liu, C. X. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880.e821 (2019).
-
Suzuki, H. et al. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34, e63 (2006).
-
Purusharth, R. I., Madhuri, B. & Ray, M. K. Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3’ end processing of 16 and 5 S ribosomal RNA. J. Biol. Chem. 282, 16267–16277 (2007).
-
Vicens, Q. & Cech, T. R. A natural ribozyme with 3’,5’ RNA ligase activity. Nat. Chem. Biol. 5, 97–99 (2009).
-
Zhang, Z. et al. Mitigating cellular dysfunction through contaminant reduction in synthetic circRNA for high-efficiency MRNA-based cell reprogramming. Adv. Sci. (Weinh.) 12, e2416629 (2025).
-
Corbett, K. S. et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020).
-
Mencin, N. et al. Development and scale-up of oligo-dT monolithic chromatographic column for mRNA capture through understanding of base-pairing interactions. Sep. Purif Technol. 304, 122320 (2023).
-
Cui, T. et al. Comprehensive studies on building a scalable downstream process for mRNAs to enable mRNA therapeutics. Biotechnol. Prog. 39, e3301 (2023).
Acknowledgements
This project was funded by the National Key Research and Development Program of China (grant no. 2022YFC0870900), the Beijing Advanced Innovation Center for Genomics at Peking University (PKU), the Peking-Tsinghua Center for Life Sciences, and Changping Laboratory (W.W.). We thank Junjie Liu (Tsinghua University) for providing the plasmids of HYER. We thank the National Center for Protein Sciences (PKU) and the Center for Quantitative Biology (PKU) for assistance with flow cytometry.
Ethics declarations
Competing interests
W.W., Y.S., and L.G. have submitted a patent application to the National Intellectual Property Administration, PRC patent office pertaining to the findings of this work (application number PCT/CN2024/094971). The applicants are Peking University, Changping Laboratary and Therorna. W.W. is a founder and scientific advisor for Therorna and EdiGene, while L.G. is employed by Therorna. The other authors report no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shen, Y., Li, B., Dong, L. et al. Self-splicing RNA circularization facilitated by intact group I and II introns. Nat Commun 16, 7376 (2025). https://doi.org/10.1038/s41467-025-62607-y
-
Received:
-
Accepted:
-
Published:
-
DOI: https://doi.org/10.1038/s41467-025-62607-y