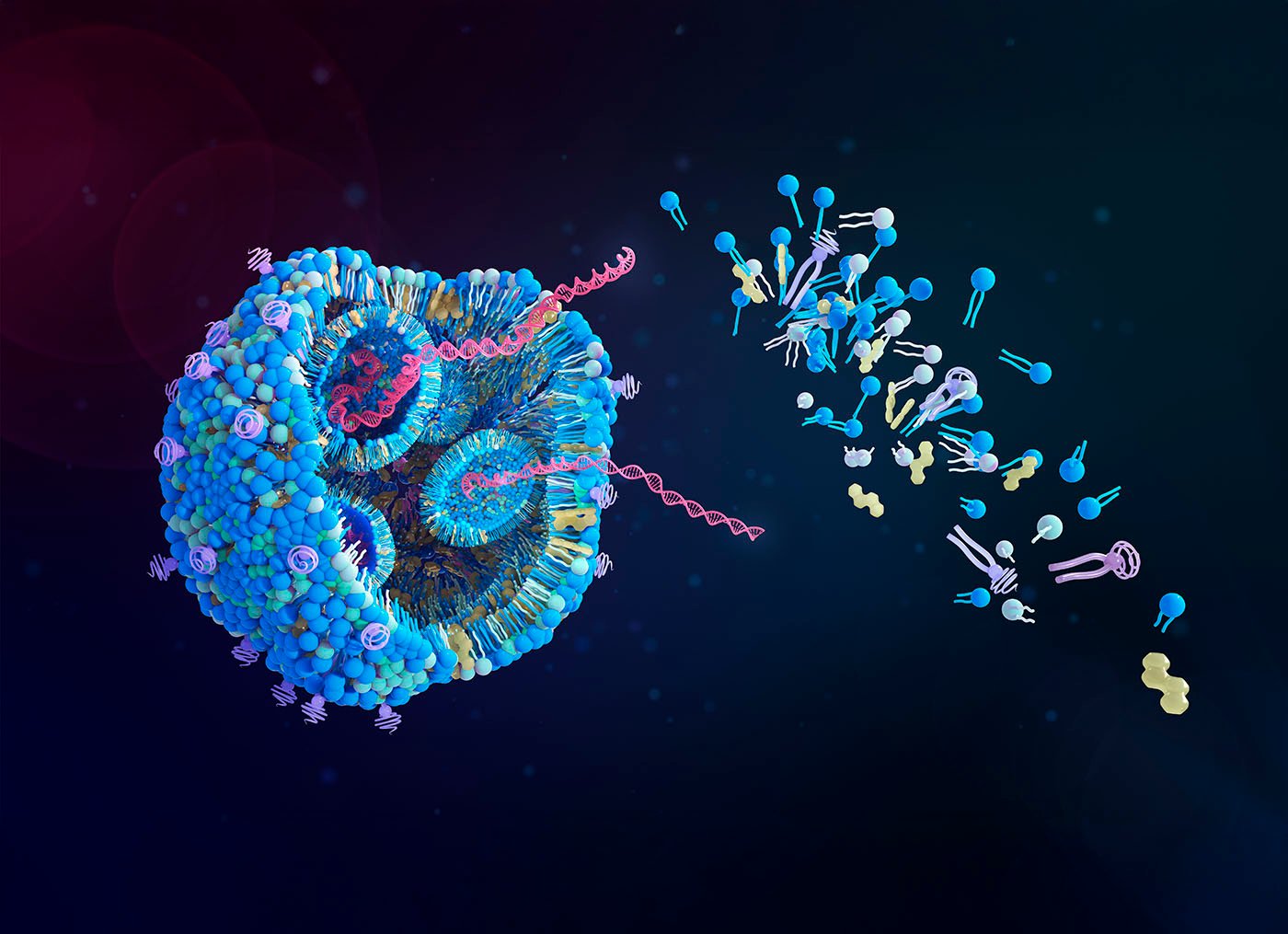
Using artificial intelligence (AI), researchers headed by a team at Massachusetts Institute of Technology (MIT) have come up with a new way to design nanoparticles that can more efficiently deliver RNA vaccines and other types of RNA therapies.
The scientists trained a machine-learning model, Composite Material Transformer (COMET) to analyze thousands of existing delivery particles, and then used this to predict new materials that would work even better. The model also enabled the researchers to identify particles that would work well in different types of cells and to discover ways to incorporate new types of materials into the particles.
“What we did was apply machine-learning tools to help accelerate the identification of optimal ingredient mixtures in lipid nanoparticles to help target a different cell type or help incorporate different materials, much faster than previously was possible,” said Giovanni Traverso, PhD, an associate professor of mechanical engineering at MIT and gastroenterologist at Brigham and Women’s Hospital.
This approach could dramatically speed the process of developing new RNA vaccines, as well as therapies that could be used to treat obesity, diabetes, and other metabolic disorders, the researchers say.
Senior author Traverso, together with lead authors Alvin Chan, PhD, a former MIT postdoc who is now an assistant professor at Nanyang Technological University, and Ameya Kirtane, PhD, a former MIT postdoc who is now an assistant professor at the University of Minnesota, described their development in Nature Nanotechnology, in a paper titled “Designing lipid nanoparticles using a transformer-based neural network.” In the paper, the team reported, “Experimental validation showed that our approach can identify LNPs that exhibit strong protein expression in vitro and in vivo, promising accelerated development of nucleic acid therapies with extensive potential across therapeutic and manufacturing applications.”
RNA vaccines, such as the vaccines for SARS-CoV-2, are usually packaged in lipid nanoparticles (LNPs) for delivery. “The RNA medicine revolution has been spurred by lipid nanoparticles (LNPs),” the team wrote. These particles protect mRNA from being broken down in the body and help it to enter cells once injected.
Creating particles that handle these jobs more efficiently could help researchers to develop even more effective vaccines. Better delivery vehicles could also make it easier to develop mRNA therapies that encode genes for proteins that could help to treat a variety of diseases. There are challenges, the researchers pointed out, “The effectiveness of an LNP is determined by its lipid components and their ratios; however, experimental optimization is laborious and does not explore the full design space.”
In 2024 Traverso’s lab launched a multiyear research program, funded by the U.S. Advanced Research Projects Agency for Health (ARPA-H), to develop new ingestible devices that could achieve oral delivery of RNA treatments and vaccines. “Part of what we’re trying to do is develop ways of producing more protein, for example, for therapeutic applications,” Traverso said. “Maximizing the efficiency is important to be able to boost how much we can have the cells produce.”
A typical LNP consists of four components—a cholesterol, a helper lipid, an ionizable lipid, and a lipid that is attached to polyethylene glycol (PEG). The authors further explained, “LNPs comprise four lipid classes, each crucial for cytosolic RNA delivery. Their function depends on lipid structures and ratios, with composition requiring re-optimization per application.”
Different variants of each of these components can be swapped in to create a huge number of possible combinations. Changing up these formulations and testing each one individually is very time-consuming, so Traverso, Chan, and colleagues decided to turn to artificial intelligence to help speed up the process.
However, Chan noted, “Most AI models in drug discovery focus on optimizing a single compound at a time, but that approach doesn’t work for lipid nanoparticles, which are made of multiple interacting components.” The authors further stated, “Computational approaches such as deep learning can be greatly beneficial, but the composite nature of LNPs limits the effectiveness of existing single molecule-based algorithms to LNPs.”
Chan continued, “To tackle this, we developed a new model called COMET, inspired by the same transformer architecture that powers large language models like ChatGPT. Just as those models understand how words combine to form meaning, COMET learns how different chemical components come together in a nanoparticle to influence its properties—like how well it can deliver RNA into cells.”
To generate training data for their machine-learning model, the researchers created the Lipid-RNA Nanoparticle Composition and Efficacy (LANCE) dataset of more than 3,000 different LNP formulations. The team tested each of these 3,000 particles in the lab to see how efficiently they could deliver their payload to cells, then fed all of this data into a machine-learning model.
“The design of COMET is motivated by the importance of not only the molecular structure of individual ingredients (for example, lipids) in drug products but also the interactions among compounds and their relative ratios,” the authors stated. “Its transformer-based architecture integrates multimodal features—including molecular structures, molar percentages and synthesis parameters—into a unified artificial intelligence framework.”
After the model was trained, the researchers asked it to predict new formulations that would work better than existing LNPs. They tested those predictions by using the new formulations to deliver mRNA encoding a fluorescent protein to mouse skin cells grown in a lab dish. They found that the LNPs predicted by the model did indeed work better than the particles in the training data, and in some cases better than LNP formulations that are used commercially.
Once the researchers showed that the model could accurately predict particles that would efficiently deliver mRNA, they began asking additional questions. First, they wondered if they could train the model on nanoparticles that incorporate a fifth component: a type of polymer known as branched poly beta amino esters (PBAEs).
Research by Traverso and his colleagues has shown that these polymers can effectively deliver nucleic acids on their own, so they wanted to explore whether adding them to LNPs could improve LNP performance. The MIT team created a set of about 300 LNPs that also include these polymers, which they used to train the model. “COMET’s flexible input format enables exploration of non-canonical formulations, such as dual-ionizable lipid LNPs or polymer–lipid hybrids (for example, branched PBAEs),” the team stated. “A dataset of 454 polymer–LNPs (13 unique PBAEs) was added to LANCE.” The resulting model could then predict additional formulations with PBAEs that would work better.
Next, the researchers set out to train the model to make predictions about LNPs that would work best in different types of cells, including a type of cell called Caco-2, which is derived from colorectal cancer cells. Again, the model was able to predict LNPs that would efficiently deliver mRNA to these cells. In addition, the researchers used the model to predict which LNPs could best withstand lyophilization—a freeze-drying process often used to extend the shelf-life of medicines. “Beyond efficacy, COMET also predicts formulation stability post-lyophilization, despite limited data,” they stated. “This accuracy improves with multitask training using LANCE. Similar gains were observed in adapting COMET to new cell types (for example, Caco-2), underscoring the broad applicability of our approach.”
Traverso added, “This is a tool that allows us to adapt it to a whole different set of questions and help accelerate development. We did a large training set that went into the model, but then you can do much more focused experiments and get outputs that are helpful on very different kinds of questions.”
The team is now working on incorporating some of these particles into potential treatments for diabetes and obesity, which are two of the primary targets of the ARPA-H funded project. Therapeutics that could be delivered using this approach include GLP-1 mimics with similar effects to Ozempic. In their paper the researchers concluded, “COMET’s architecture may also support links to other areas of nanotechnology where multi-component formulations are critical, such as co-delivery of multiple cargos, immunomodulatory nanoparticle design, or materials for tissue engineering.”
The post COMET’s Rocket Speed: AI-Designed Nanoparticles Accelerate mRNA Therapies appeared first on GEN – Genetic Engineering and Biotechnology News.