Introduction
Unlike cigarettes, cigars are made of whole tobacco leaves. Before rolling into cigars, tobacco leaves undergo air-curing, fermentation, and ageing. Fermentation is the most critical process that accounts for the development of rich flavour and aroma, as well as changes in surface integrity1,2. To ignite fermentation, air-cured cigar tobacco leaves are rehydrated and piled up. Under the pressure of their weight, leaves slowly heat up and are unravelled when the central temperature reaches 35 °C. This is an intense yet closely controlled process in which microbial activity is the driving force of the physical and chemical transition3. Macromolecules in tobacco leaves such as sugars, proteins, carotenoids, and alkaloids are broken down by microorganisms and enzymes into smaller molecules, including reducing sugars, organic acids, amino acids, ammonia, and other flavour compounds4. Simultaneously, synthetic reactions (i.e. Maillard reaction, caramelisation, and esterification reaction) take place and create a roasted, nutty aroma5,6. As a result, fermentation achieves balanced organoleptic qualities by eliminating the irritation, bitterness, and impurities in unpalatable raw tobacco leaves, as well as developing nuanced aromas and flavours.
Due to the unique microclimate and soil, tropical regions are ideal for the growth of premium cigar tobacco, among which countries in Central America such as Cuba are known for top-notch cigars. Apart from environmental conditions, microorganisms are exceedingly crucial to the quality and flavour profile of cigar tobacco leaves by participating in the fermentation process. Tobacco leaf-associated microorganisms are mainly derived from the environment, and hence, distinctive microbial structures and volatile flavour compounds have been observed among tobacco leaves sourced from different regions3,7,8. Although fungi and yeasts are involved in the fermentation process through saprophytic activity, bacteria not only have higher diversity and abundance but also play a greater role in flavour formation3. Lately, with the development of high-throughput sequencing, several studies have focused on the dominant microbial species and their correlation with metabolites in cigar tobacco leaves4,9. Bacteria including Bacillus, Staphylococcus, Pseudomonas, Corynebacterium, Aerococcus, and Lactobacillus are commonly found as dominant species in tobacco leaves3,10,11. Among them, Bacillus, Vibrio, Sphingomonas, Thioalkalicoccus, and Jeotgalicoccus are positively correlated with volatile flavour compounds3,11. Therefore, with the purpose of either accelerating the fermentation rate or manipulating tobacco organoleptic qualities, certain microorganisms have been artificially added to the fermentation medium. As early as 1858, Koller added yeast to cigar tobacco leaves12. Since then, various microbial species isolated from tobacco leaves have been supplemented into the fermentation medium, including Bacillus13, Acinetobacter14, and Candida15. The outcome of these artificial fermentation approaches includes increased content of volatile flavour compounds, altered tobacco quality, and shortened fermentation period.
In the past decade, China’s cigar market has experienced a surge, and meanwhile, domestic cultivation and production of cigar tobacco have expanded enormously4. Nevertheless, the cigar industry is restricted by the inferior quality of domestic cigar raw materials and is heavily reliant on imports. Thus far, previous multi-omics studies mainly revolved around either microbial and chemical changes of cigar tobacco leaves during the fermentation/ageing process and their correlation with sensory quality2,16; or bacterial communities of machine-made cigars (usually containing homogenised tobacco leaves) instead of hand-rolled cigars17,18,19. However, to our best knowledge, microbial composition in finished cigars, especially premium Cuban-made cigars has never been studied. Hence, this study aimed to compare microbial communities and organoleptic qualities between Caribbean cigars and Chinese cigars, by high-throughput sequencing and sensory evaluation. It sheds light on the roles of microbial communities during fermentation and the development of microbial agents for bioaugmentation fermentation.
Materials and methods
Sample collection
Three Cuban cigar products (sample M, RB, and RP), two Dominican cigar products (sample CE and FH), and five Chinese cigar products (sample A, DF, DS, JY, and MZ) were acquired from the Quality Supervision and Test Center, China National Tobacco Corporation Shandong Branch. Cigars were kept at 20 °C with a 72% relative humidity in their original factory packages.
Sequencing method
To avoid contamination, tools in contact with samples were autoclaved and treated with UV beforehand, and containers of DNA-free level were used. In a Class 2 biosafety cabinet, a cigar sample (5 g) was cut using a sterile blade and ground into powder with liquid nitrogen in a sterile mortar. Total genome DNA from samples was extracted using the cetyltrimethylammonium bromide method. DNA yield and purity were monitored on 1% agarose gels.
The V3-V4 region of the 16S rRNA gene was amplified using the 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) primer pair with specific barcodes. All PCR reactions were carried out in a final volume of 30 μl, containing 0.2 μmol/L primer pair, 10 ng template DNA, and 15 μl Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, US). Thermal cycling consisted of initial denaturation at 98˚C for 1 min, followed by 30 cycles of denaturation at 98˚C for 10 secs, annealing at 50˚C for 30 s, and elongation at 72˚C for 30 s. Finally, one cycle at 72˚C for 5 min.
PCR products were mixed with the same volume of loading buffer (containing SYBR-Green) and analysed on a 2% agarose gel. Samples with a bright main strip between 400 and 450 bp were chosen for further experiments.
Sequencing libraries were generated using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina (New England Biolabs) and index codes were added. Library quality was assessed on a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, US) and an Agilent Bioanalyzer 2100 system. At last, the library was sequenced on a NovaSeq platform (Illumina, San Diego, CA, US) and 250 bp paired-end reads were generated.
Sensory quality evaluation
Sensory quality evaluation was carried out by the Quality Supervision and Test Center, China National Tobacco Corporation Shandong Branch. By the time of sensory evaluation, there were not enough cigar samples for CE. Therefore, four Caribbean cigar products and five Chinese cigar products were evaluated by five qualified experts according to the national standard “Cigars—Part 4: Technical Requirements for Sense Evaluation” (GB 15,269.4–2011). The sensory quality of cigars was scored for seven indices including colour (0–6 points), oily (0–4 points), flavour (0–36 points), offensive taste (0–12 points), irritancy (0–14 points), after taste (0–22 points), and ashing degree (0–6 points).
Data analysis
Paired-end reads were merged by FLASH (version 1.2.11, http://ccb.jhu.edu/software/FLASH/), processed using fastp (version 0.20.0) to perform quality filtering, and Vsearch (Version 2.15.0) software to obtain Effective Tags. Amplicon sequence data were analysed using QIIME software (QIIME2-2020.6), and qiime diversity alpha and qiime diversity core-metrics-phylogenetic were used to analyse α (within samples) and β (among samples) diversity metrics, respectively. First, denoise was performed with DADA2 to obtain Amplicon Sequence Variants, with abundance lower than 5 filtered out. Then, species annotation and phylogenetic relationship construction were performed using QIIME2 software. The absolute abundance of Amplicon Sequence Variants was normalized using a standard of sequence number corresponding to the sample with the fewest sequences. Subsequent analysis of α diversity and β diversity was all performed based on the output normalized data.
In order to analyse the diversity, richness, and uniformity of the communities in the sample, α diversity was calculated from 4 indices in QIIME2, including ACE, Chao1, Shannon, and Simpson. The Kruskal–Wallis test was used to determine the statistical significance of α diversity indices in QIIME2. Rarefaction curves were generated based on Observed Species to demonstrate sequencing depth. QIIME2 calculates both weighted and unweighted UniFrac, which are phylogenetic measures of β diversity. Principal coordinate analysis (PCoA) was analysed based on the Bray–Curtis distance, an indicator used to measure differences in taxonomic composition in ecology. To study the significance of the differences in community structure between groups, the Adonis and Anosim functions in QIIME2 were used to do the analysis. To find out the significantly different species at each taxonomic level (Phylum, Class, Order, Family, Genus, Species), R (version 4.2.2) was used to do MetaStat and T-test analysis. The LEfSe package in R was used to do LEfSe analysis (linear discriminant analysis effect size) with a linear discriminant analysis (LDA) score threshold of 4 to find out the biomarkers. Further, to study the functions of the communities in the samples and find out the different functions of the communities in the different groups, the PICRUSt2 software (Version 2.1.2-b) was used for function annotation analysis.
To discern differences among cigar-associated groups at the genus and species levels, a comparative analysis was conducted. The ‘pheatmap’ package (version 1.0.13) in R (version 4.2.2) was utilised to generate heatmaps, while the ‘ggplot2’ (version 3.5.2) R package was employed to create histograms illustrating the average relative abundance within each group. Correlation analysis was performed at the genus and species levels. Genera and species identified in the variation analysis were pooled for Spearman correlation analysis of their correlation with all the bacteria and clinical parameters after 0.002% and 30% screening (R package: ‘Hmisc’, version 5.2–3, Supplement Information).
Results
Characteristics of microbial communities in cigar samples
Cigar samples of five Caribbean products and five Chinese products were subjected to 16S rRNA sequencing analysis in replicates of six. Splicing, quality control, and chimera removal were conducted on the down data acquired from Illumina NovaSeq to obtain Effective Tags. The statistical results obtained in each step of data processing are detailed in Table S1.
As shown in Tables S1 and S2, bacterial sequencing revealed that Caribbean and Chinese cigars yielded 3,249,417 and 3,182,671 Effective Tags in total, which were classified into 3026 operational taxonomic units and 12,199 OTUs, respectively. Rarefaction curves were created by randomly selecting a subset of sequencing data from the sample and plotting against observed OTUs. In Fig. S1, rarefaction curves showed that the number of OTUs in all samples reached a plateau as the sequence number increased. This suggests that the OTUs identified in our data represent sufficient coverage of the microbial diversity present in the samples.
The microbiome structure showed that Caribbean and Chinese cigars had distinct α and β diversity
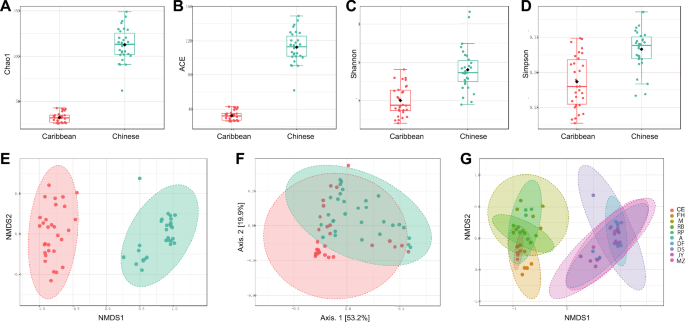
Microbial diversities were analysed to explore the differences in microbial communities of cigars produced from different countries. The Chao1 and ACE indices, which estimate community richness, indicated significant differences in within-sample diversity between the Caribbean and Chinese cigars (Fig. 1a,b , p<0.0001). This finding was further supported by Shannon and Simpson indices, which assess community diversity, respectively (Fig. 1c,d, p<0.0001). However, there was no significant difference in within-sample diversity among Caribbean cigars or among Chinese cigars (Fig. S2).
Diagrams of α and β diversity analysis of grouped cigar samples. Indices of α diversity, (a) Chao 1 index, (b) ACE index, (c) Shannon index, (d) Simpson index. Indices of β diversity, (e) NMDS and (f) PCoA between Caribbean and Chinese cigar products, (g) NMDS analysis of within-sample diversity among each product of Caribbean and Chinese cigar products. In a to f, coral color represents Caribbean cigars, while turquoise color represents Chinese cigars.
Beta diversity analysis revealed significant differences in the between-sample diversity of Caribbean and Chinese cigars. Compared to Caribbean cigars, Chinese cigars exhibited higher between-sample microbial diversity, as evidenced by higher values in both Non-metric Multidimensional Scaling (NMDS) and PCoA (Fig. 1e,f, p< 0.05). Similarly, β diversity exhibited no significant variations either across the spectrum of Caribbean cigar samples or within the realm of Chinese domestic cigars (Fig. 1g). These results suggested that Chinese cigars harboured more diverse microbial communities compared to Caribbean cigars, supported by higher α and β diversity.
The comparison of the core microbiome between Caribbean and Chinese cigars
A thorough examination of the microbial composition within the samples was conducted, focusing on the phylum and genus levels. At the phylum level, bacterial communities in all samples were predominantly composed of Cyanobacteria, Bacillota, Proteobacteria, and Actinobacteria, which accounted for over 95% of the total population (Fig. S3). Specifically, Cyanobacteria and Bacillota were the most and second most abundant phyla, respectively. However, there was a notable difference in the distribution of these phyla between Caribbean and Chinese cigars. In Caribbean cigars, Bacillota was the dominant phylum, constituting more than half of the total bacterial count and exceeding 90% in some samples. In contrast, its prevalence in Chinese cigars was generally less than 50%. Conversely, the distribution of Cyanobacteria showed a significant similarity between the two groups (Fig. S3). At the genus level, the microbiome structure was similar to that at the phylum level. The genus Staphylococcus, a member of the Bacillota phylum, was the most abundant and accounted for more than half of the total population in Caribbean cigars (Fig. 2a). In Chinese cigars, however, Staphylococcus constituted only about 30% of the bacterial population in most samples (Fig. 2b).
Comparisons of the core microbiota between Caribbean and Chinese cigars. Stacked bar graphs representing the relative abundance of the top 10 genera across Caribbean (a) and Chinese cigars (b). Venn diagrams show the number of unique and shared genera in the core microbiome of each product of Caribbean cigars (c) and Chinese cigars (d), as well as Caribbean vs. Chinese cigars (e).
Next, Venn analysis was employed to identify the core microbiome shared among the five cigars within the Caribbean and Chinese cigars. The analysis at the genus level revealed that the core microbiome of Chinese cigars encompassed a greater number of bacterial genera compared to that of Caribbean counterparts (316 genera vs. 51 genera, Fig. 2c,d). Among them, 184 out of 316 genera of Chinese cigars had definite genera, while the number was 33 out of 51 for Caribbean cigars. Among the definite genera constituting the core microbiome of both groups, 21 genera were found to be shared (Fig. 2e, and Table S3). The core microbiome of Caribbean cigars uniquely included six genera, which comprised three genera from the phylum Proteobacteria (Haemophilus, Neisseria, and Serratia), three genera from the phylum Bacillota (Brochothrix, Lactococcus, and Carnobacterium). These findings underscored the differences in the core microbiome between Chinese and Caribbean cigars. The unique genera present in the core microbiome of Caribbean cigars may be a key factor that contributes to their quality.
Differences in the microbial communities of Caribbean and Chinese cigars
LEfSe analysis was applied to discover high-dimensional biomarkers with statistical and biological significance, and the results included cladograms and LDA value distribution bar graphs. Cladograms provided a comprehensive overview of the microbial structure of cigars, highlighting the primary bacterial differences. In the genera with relatively high abundance, Staphylococcus, Ralstonia, Brochothrix, Weissella and Carnobacterium were notably enriched in the microbiome of Caribbean cigars, whereas Acinetobacter, Aureimonas, Bacillus, Streptococcus, Enteractinococcus, Brevibacterium, Atopostipes, Pseudoalteromonas, Terribacillus, Lactobacillus, and Petrimonas were predominant in Chinese cigars (Fig. 3a,c). Except for Petrimonas, all these bacteria were aerobic. Moreover, the top five differential genera in the microbiome of cigars of different origins revealed a higher representation of Bacillota in Caribbean cigars and a greater abundance of Proteobacteria in Chinese cigars (Fig. S3), indicating a distinct difference in the distribution of these phyla between cigars from different origins. Upon comparing the microbiome of all cigars, the genera enriched in each sample were as follows: CE (Staphylococcus, Weissella, and Brochothrix), FH (Corynebacterium, Aerococcus, Facklamia, and Oceanobacillus), M (Chloroplast), RB (Tetragenococcus and Carnobacterium), RP (Ralstonia and Prevotella), A (Carnimonas, Mannheimia, Mitochondria, and Streptococcus), DF (Klebsiella, Pasteurella, Atopococcus, and Escherichia-Shigella), DS (Sphingomonas, Aureimonas, Atopobium, and Brevibacterium), and JY (Enteractinococcus, Fastidiosipila, Petrimonas, and Lentibacillus), MZ (Atopostipes and Shimwellia) (Fig. 3b,d). These findings suggested that the key microbiome of Caribbean and Chinese cigars was distinct, with each product exhibiting its characteristic bacterial genera.
Statistical results of LEfSe analysis. In cladograms (a for comparison between Caribbean and Chinese, and b for comparison among each product), the circles radiating from the inside to the outside represent the classification level from class to genus. Each small circle at a different classification level represents a classification at that level, and the diameter of the small circle is proportional to the relative abundance. Colouring principle: biomarkers with no significant differences were uniformly in white, and the biomarkers were coloured according to the group. LDA value distribution bar graphs (c for Caribbean vs. Chinese and d for comparison among each product) show biomarkers with statistical differences between groups (LDA scores ≥ 4). The length of the bar chart represents the impact size of each taxon.
Variations in cigar microbiome at the functional level
The phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) method was applied to predict possible metabolic activities of the microbial communities present in cigars at the functional level, comparing those between Caribbean and Chinese cigars. The study identified core pathways, which are metabolic functions common to a specific group of samples, and found that Chinese cigars exhibited a greater number of these pathways compared to those for Caribbean cigars (398 versus 355, as depicted in Fig. 4a,b, and Table S4). The top 35 signalling pathways of the microbial communities harboured by the two sets of cigars are shown in Fig. 4c,d. Among these core pathways, 354 were shared by both groups (Fig. 4e). The pathway exclusive to the core pathways of Caribbean cigars was PWY-6148, which is associated with the biosynthesis of tetrahydromethanopterin. This signalling pathway consists of 11 enzymes, and bacteria encoding at least one enzyme include Acinetobacter, Anaplasma, Bartonella, Burkholderia, Ehrlichia, Helicobacter, Mycobacterium, and Pseudomonas. Notably, Acinetobacter was identified as one of the differentially abundant bacteria within the core microbiome, and it was among the taxa with the highest abundance. These functional variations suggested the potential impact of microbial communities on the physicochemical properties of cigar tobacco leaves.
Functional differences of microbial communities on cigars using PICRUSt2. Venn diagrams presenting unique and shared core pathways in microbial communities of Caribbean (a) and Chinese (b) cigar products. Clustered heatmaps of the relative abundance of the top 35 pathways in Caribbean cigars (c) and Chinese cigars (d). On the left side of heatmaps is the functional clustering tree. The value corresponding to each block was the Z value of each row of the pathway, representing the standardized relative abundance. (e) Venn diagram showing unique and shared core pathways among cigars from different regions.
The correlation between microbial community and sensory quality of cigars
Correlation analysis between microbial communities and sensory quality in the Caribbean and Chinese cigars has provided significant insights. The result of the sensory evaluation is listed in Table S5. Spearman correlation analysis revealed a global correlation between attributes of sensory evaluation and the microbial communities (Fig. 5a). Specifically, the genera Weissella and Neisseria exhibited a positive association with flavour, whereas Amaricoccus and Tumebacillus showed a negative association. In terms of offensive taste, the genus Desemzia was positively correlated, while Rhodococcus was negatively correlated. Regarding irritancy, Ornithinimicrobium was positively linked, while Clostridium sensu stricto 13 bacteria were negatively linked. Finally, the Petrimonas genus was positively correlated with ashing degree, while Atopostipes was negatively correlated.
The correlation between microbial communities and sensory quality using Spearman correlation analysis. The correlation in Caribbean and Chinese cigars combined (a), Caribbean cigars (b), and Chinese cigars (c). A p-value less than 0.05 was regarded as statistically significant and labelled with an asterisk.
Figure 5b,c show the correlation between microbial communities and sensory qualities in the group of Caribbean and Chinese cigars, respectively. In Caribbean cigars, Brachybacterium, Corynebacterium, Staphylococcus, Atopococcus, Methylobacterium-Methylorubrum, Ornithinimicrobium, Mitochondria, Neisseria, Aerococcus, Facklamia, and Sphingomonas were associated with certain attributes of sensory quality, while Atopococcus, Clostridium sensu stricto 13, Amaricoccus, Escherichia-Shigella, Uruburuella, Mesorhizobium, Carnimonas, Haemophilus, Mannheimia, Natribacillus, Pseudoalteromonas, Lawsonella, and SJA-28 were associated with certain attributes in Chinese cigars. Notably, Staphylococcus positively correlated with the sum in Caribbean cigars, while Amaricoccus, Escherichia-Shigella, and Uruburuella were linked to the sum in Chinese cigars. These findings suggested that certain bacteria could play a pivotal role in the appearance and chemical composition of cigar tobacco leaves, and therefore affect the sensory quality of cigars.
Discussion
In the fermentation and ageing process, cigar tobacco leaves go through an intense transformation, where microorganisms play a critical role by manipulating the degradation of macromolecules11. Metabolic byproducts of microorganisms, such as organic acids, ketones, and aldehydes, significantly impact the olfactory and gustatory profiles of tobacco leaves11. Certain microbes, for instance, Pseudomonas and Pseudomonadales reduce the level of nicotine in tobacco, mitigating irritation of the smoke20. Modification of the microbial consortia could indirectly yet significantly, affect the intrinsic quality of tobacco leaves15. However, studies aimed at elucidating the function of microorganisms of cigar tobacco leaves through mining microbial composition and sensory quality remain unclear4,9. Consequently, this study conducted a comparative analysis of the microbial profiles of Caribbean- and Chinese-produced cigars, revealing significant differences in the composition of cigar microbiomes from different regions, potentially accounting for the variations in their quality and flavour. To our knowledge, this is the first study to investigate the microbial composition of premium Cuban cigars.
Our findings indicated that Chinese cigars harboured a more diverse array of microorganisms compared to Caribbean counterparts, as evidenced by indices of α and β diversity. This is not consistent with previous reports on cigar tobacco leaves, where either Caribbean cigars showed higher or similar diversity than Chinese cigars3,7. The composition of the microbiome revealed that the genus Staphylococcus, belonging to the Bacillota phylum, was predominant in Caribbean cigars, representing over half of the total bacterial population. In contrast, Staphylococcus merely made up approximately 30% of the bacterial community in most Chinese cigar samples. Staphylococcus shows high abundance through all stages of fermentation and is associated with the degradation of starch, lipids, and amino acids1,15,21. Among the genera with higher abundance, Staphylococcus, Ralstonia, Brochothrix, Weissella, and Carnobacterium were significantly more prevalent in the microbiome of Caribbean cigars. On the other hand, Acinetobacter, Aureimonas, Bacillus, Streptococcus, Enteractinococcus, Brevibacterium, Atopostipes, Pseudoalteromonas, Terribacillus, Lactobacillus, and Petrimonas were more commonly found in Chinese cigars. The microbial disparities between Caribbean and Chinese cigars implied potential chemical alterations caused by microbial communities. Subsequent comprehensive statistical analysis of the correlation between microbiome and sensory quality of Caribbean and Chinese cigars suggested that certain bacterial species might be fundamental in the sensory quality of cigars, especially flavour and aroma. These insights indicated that the dominant bacteria of Caribbean and Chinese cigars were highly distinct, with each product exhibiting its characteristic bacterial genera. The genera exclusive to Caribbean cigars might be applied to fermentation and ageing processes, leading to alterations in flavour and aroma profiles. This knowledge could lay the foundation for artificial fermentation by supplementing fermentation liquid with certain microorganisms14,15. Bacterial strains isolated from samples in this study have been subjected to further genome sequencing and enzymatic activity examination22. Nevertheless, as microorganisms engage in complex interactions, bioaugmentation fermentation with only a few microbial isolates might not achieve desirable effects. Developing and applying novel biotechnologies, including enzymes and microbial consortia fermentation starter, could promote biochemical reactions during the fermentation process. Multi-omics techniques such as genomics, metabolomics, and proteomics are utilised for the in-depth study of microbial community structure and function21. In this study, we highlighted some characteristic bacteria in premium Caribbean cigars and discussed their unique metabolic processes.
While this study revealed the composition and function of microorganisms in cigar tobacco, it has certain limitations. Since microbial composition fluctuates greatly during the fermentation and ageing process, high-throughput sequencing performed on the finished cigar products might not reflect key species that shape the quality of cigar tobacco leaves2. Nevertheless, this is restricted by the embargo on cigar tobacco leaves implemented by Cuba in the 1980s. In addition, a correlation analysis between chemical compounds (i.e. sugars, alkaloids, and aromatic compounds) and microbial compositions would provide a better understanding of the function of microbial communities.
In summary, the insights into the microbiome of cigars from this research could pave the way for advancing artificial fermentation technologies. These could be instrumental in changing the intrinsic quality of cigar tobacco leaves, including their aroma, flavour, and combustion characteristics—a key focus of research in the Chinese tobacco industry. Further research is warranted to elucidate the nexus between microbial communities and sensory quality, as well as the application of key microorganisms in the bioaugmentation fermentation of Chinese domestic cigars.