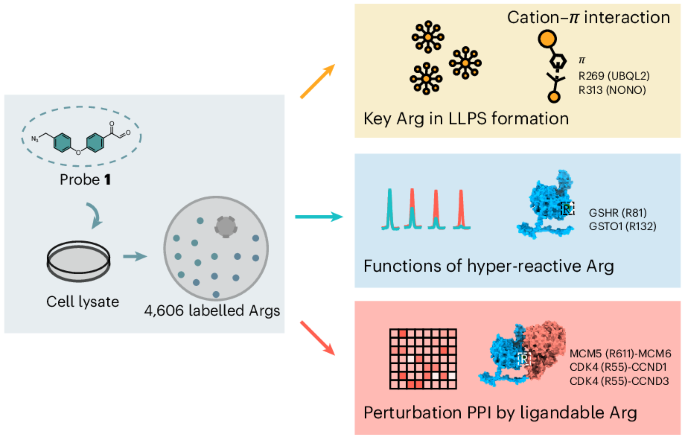
Jin, Y., Jana, S., Abbasov, M. E. & Lin, H. Antibiotic target discovery by integrated phenotypic and activity-based profiling of electrophilic fragments. Cell Chem. Biol. 32, 434–448.e9 (2025).
Zhang, X. & Cravatt, B. F. Chemical proteomics–guided discovery of covalent ligands for cancer proteins. Annu. Rev. Cancer Biol. 8, 155–175 (2024).
Niphakis, M. J. & Cravatt, B. F. Ligand discovery by activity-based protein profiling. Cell Chem. Biol. 31, 1636–1651 (2024).
Offensperger, F. et al. Large-scale chemoproteomics expedites ligand discovery and predicts ligand behavior in cells. Science 384, eadk5864 (2024).
Liu, Y., Patricelli, M. P. & Cravatt, B. F. Activity-based protein profiling: the serine hydrolases. Proc. Natl Acad. Sci. USA 96, 14694–14699 (1999).
Weerapana, E. et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010).
Shi, Y., Fu, L., Yang, J. & Carroll, K. S. Wittig reagents for chemoselective sulfenic acid ligation enables global site stoichiometry analysis and redox-controlled mitochondrial targeting. Nat. Chem. 13, 1140–1150 (2021).
Fu, L. et al. Nucleophilic covalent ligand discovery for the cysteine redoxome. Nat. Chem. Biol. 19, 1309–1319 (2023).
Qin, W. et al. S-glycosylation-based cysteine profiling reveals regulation of glycolysis by itaconate. Nat. Chem. Biol. 15, 983–991 (2019).
Hacker, S. M. et al. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 9, 1181–1190 (2017).
Abbasov, M. E. et al. A proteome-wide atlas of lysine-reactive chemistry. Nat. Chem. 13, 1081–1092 (2021).
Hahm, H. S. et al. Global targeting of functional tyrosines using sulfur-triazole exchange chemistry. Nat. Chem. Biol. 16, 150–159 (2020).
Sun, F., Suttapitugsakul, S. & Wu, R. An azo coupling-based chemoproteomic approach to systematically profile the tyrosine reactivity in the human proteome. Anal. Chem. 93, 10334–10342 (2021).
Chen, Y. et al. Direct mapping of ligandable tyrosines and lysines in cells with chiral sulfonyl fluoride probes. Nat. Chem. 15, 1616–1625 (2023).
Lin, S. X. et al. Redox-based reagents for chemoselective methionine bioconjugation. Science 355, 597–602 (2017).
Bach, K., Beerkens, B. L. H., Zanon, P. R. A. & Hacker, S. M. Light-activatable, 2,5-disubstituted tetrazoles for the proteome-wide profiling of aspartates and glutamates in living bacteria. ACS Cent. Sci. 6, 546–554 (2020).
Ma, N. et al. 2H-Azirine-based reagents for chemoselective bioconjugation at carboxyl residues inside live cells. J. Am. Chem. Soc. 142, 6051–6059 (2020).
Xie, X. et al. Oxidative cyclization reagents reveal tryptophan cation–π interactions. Nature 627, 680–687 (2024).
Zhai, Y. et al. Global profiling of functional histidines in live cells using small-molecule photosensitizer and chemical probe relay labelling. Nat. Chem. 16, 1546–1557 (2024).
Sharma, H. A. et al. Proteomic ligandability maps of phosphorus(V) stereoprobes identify covalent TLCD1 inhibitors. J. Am. Chem. Soc. 147, 15554–15566 (2025).
Backus, K. M. et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016).
Wang, Q. et al. Quantitative chemoproteomics reveals dopamine’s protective modification of Tau. Nat. Chem. Biol. 21, 1341–1350 (2025).
Wang, C., Weerapana, E., Blewett, M. M. & Cravatt, B. F. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 11, 79–85 (2014).
Hodges, A. J. et al. Histone sprocket arginine residues are important for gene expression, DNA repair and cell viability in Saccharomyces cerevisiae. Genetics 200, 795–806 (2015).
Bogan, A. A. & Thorn, K. S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998).
Bartlett, G. J., Porter, C. T., Borkakoti, N. & Thornton, J. M. Analysis of catalytic residues in enzyme active sites. J. Mol. Biol. 324, 105–121 (2002).
Gupta, M. N. & Uversky, V. N. Biological importance of arginine: a comprehensive review of the roles in structure, disorder, and functionality of peptides and proteins. Int. J. Biol. Macromol. 257, 128646 (2024).
Fuhrmann, J., Clancy, K. W. & Thompson, P. R. Chemical biology of protein arginine modifications in epigenetic regulation. Chem. Rev. 115, 5413–5461 (2015).
Qamar, S. et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734.e15 (2018).
Hofweber, M. et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13 (2018).
Hellwig, M. & Henle, T. Baking, ageing, diabetes: a short history of the Maillard reaction. Angew. Chem. Int. Ed. 53, 10316–10329 (2014).
Takahashi, K. The reaction of phenylglyoxal with arginine residues in proteins. J. Biol. Chem. 243, 6171–6179 (1968).
Dovgan, I. et al. Arginine-selective bioconjugation with 4-azidophenyl glyoxal: application to the single and dual functionalisation of native antibodies. Org. Biomol. Chem. 16, 1305–1311 (2018).
Jones, A. X. et al. Improving mass spectrometry analysis of protein structures with arginine-selective chemical cross-linkers. Nat. Commun. 10, 3911 (2019).
Lewallen, D. M. et al. Chemical proteomic platform to identify citrullinated proteins. ACS Chem. Biol. 10, 2520–2528 (2015).
Bicker, K. L., Subramanian, V., Chumanevich, A. A., Hofseth, L. J. & Thompson, P. R. Seeing citrulline: development of a phenylglyoxal-based probe to visualize protein citrullination. J. Am. Chem. Soc. 134, 17015–17018 (2012).
Zanon, P. R. A. et al. Profiling the proteome-wide selectivity of diverse electrophiles. Nat. Chem. 17, 1712–1721 (2025).
Wang, Q. et al. Global profiling of arginine dimethylation in regulating protein phase separation by a steric effect-based chemical-enrichment method. Proc. Natl Acad. Sci. USA 119, e2205255119 (2022).
Liu, Z., Wang, K. & Ye, M. Photoreactive probe-based strategy enables the specific identification of the transient substrates of methyltransferase at the proteome scale. Anal. Chem. 95, 12580–12585 (2023).
Shi, Y. et al. Enabling global analysis of protein citrullination via biotin thiol tag-assisted mass spectrometry. Anal. Chem. 94, 17895–17903 (2022).
Chen, P. et al. Cell-active, arginine-targeting irreversible covalent inhibitors for non-kinases and kinases. Angew. Chem. Int. Ed. 64, e202422372 (2025).
Zhang, Z., Morstein, J., Ecker, A. K., Guiley, K. Z. & Shokat, K. M. Chemoselective covalent modification of K-Ras(G12R) with a small molecule electrophile. J. Am. Chem. Soc. 144, 15916–15921 (2022).
Prosser, L., Emenike, B., Sihag, P., Shirke, R. & Raj, M. Chemical carbonylation of arginine in peptides and proteins. J. Am. Chem. Soc. 147, 10139–10150 (2025).
Kong, A. T., Leprevost, F. V., Avtonomov, D. M., Mellacheruvu, D. & Nesvizhskii, A. I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520 (2017).
Sletten, E. M. & Bertozzi, C. R. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 48, 6974–6998 (2009).
Weerapana, E., Speers, A. E. & Cravatt, B. F. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)—a general method for mapping sites of probe modification in proteomes. Nat. Protoc. 2, 1414–1425 (2007).
Chang, J. W., Lee, G., Coukos, J. S. & Moellering, R. E. Profiling reactive metabolites via chemical trapping and targeted mass spectrometry. Anal. Chem. 88, 6658–6661 (2016).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2021).
Cheng, J. et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 381, eadg7492 (2023).
Boersema, P. J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A. J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Yang, F., Gao, J., Che, J., Jia, G. & Wang, C. A dimethyl-labeling-based strategy for site-specifically quantitative chemical proteomics. Anal. Chem. 90, 9576–9582 (2018).
You, K. et al. PhaSepDB: a database of liquid-liquid phase separation related proteins. Nucleic Acids Res. 48, D354–D359 (2019).
Bryant, P., Pozzati, G. & Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 13, 1265 (2022).
Burke, D. F. et al. Towards a structurally resolved human protein interaction network. Nat. Struct. Mol. Biol. 30, 216–225 (2023).
Dao, T. P. et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978.e6 (2018).
Yamazaki, T. et al. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053.e7 (2018).
Yang, F., Jia, G., Guo, J., Liu, Y. & Wang, C. Quantitative chemoproteomic profiling with data-independent acquisition-based mass spectrometry. J. Am. Chem. Soc. 144, 901–911 (2022).
Berkholz, D. S., Faber, H. R., Savvides, S. N. & Karplus, P. A. Catalytic cycle of human glutathione reductase near 1 Å resolution. J. Mol. Biol. 382, 371–384 (2008).
Zhang, R. et al. Pleiotropic effects of a mitochondrion-targeted glutathione reductase inhibitor on restraining tumor cells. Eur. J. Med. Chem. 248, 115069 (2023).
Burger, N. et al. The human zinc-binding cysteine proteome. Cell 188, 832–850.e27 (2025).
Board, P. G. et al. S-(4-nitrophenacyl)glutathione is a specific substrate for glutathione transferase omega 1-1. Anal. Biochem. 374, 25–30 (2008).
Ramkumar, K. et al. Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor. Nat. Commun. 7, 13084 (2016).
Liu, Y. et al. Proteome-wide ligand and target discovery by using strain-enabled cyclopropane electrophiles. J. Am. Chem. Soc. 146, 20823–20836 (2024).
Baltgalvis, K. A. et al. Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase. Nature 629, 435–442 (2024).
Uechi, H. et al. Small-molecule dissolution of stress granules by redox modulation benefits ALS models. Nat. Chem. Biol. 21, 1577–1588 (2025).
Tanikawa, C. et al. Citrullination of RGG motifs in FET proteins by PAD4 regulates protein aggregation and ALS susceptibility. Cell Rep. 22, 1473–1483 (2018).
Hofweber, M. & Dormann, D. Friend or foe—post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294, 7137–7150 (2019).
Communi, D., Lecocq, R. & Erneux, C. Arginine 343 and 350 are two active site residues involved in substrate binding by human type I D-myo-inositol 1,4,5-trisphosphate 5-phosphatase. J. Biol. Chem. 271, 11676–11683 (1996).
Chen, G. & Chen, X. Arginine residues in the active site of human phenol sulfotransferase (SULT1A1). J. Biol. Chem. 278, 36358–36364 (2003).
Yan, J., He, G., Yan, F., Zhang, J. & Zhang, G. The dicarbonylation of indoles via Friedel–Crafts reaction with dicarbonyl nitrile generated in situ and retro-cyanohydrination. RSC Adv. 6, 44029–44033 (2016).
Hirapara, P. et al. CO2-assisted synthesis of non-symmetric α-diketones directly from aldehydes via C–C bond formation. Green Chem. 19, 5356–5360 (2017).
Li, J. et al. ACR-based probe for the quantitative profiling of histidine reactivity in the human proteome. J. Am. Chem. Soc. 145, 5252–5260 (2023).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217 (2019).
Wang, Y. Global profiling of arginine reactivity and ligandability in the human proteome. Zenodo https://doi.org/10.5281/zenodo.15493178 (2025).