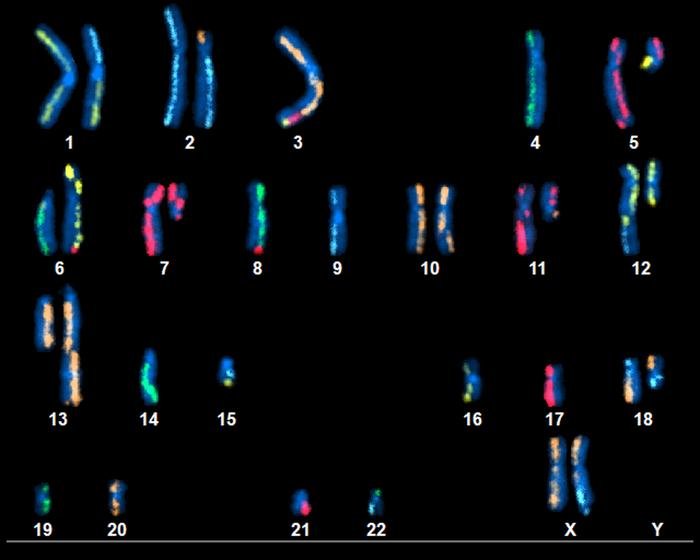
In an effort to foster progress in cancer research, the National Institute of Standards and Technology (NIST) is releasing detailed and comprehensive data about the entire genetic content of a pancreatic cancer cell.
The genome came from a 61-year-old pancreatic cancer patient who explicitly consented to making the genetic code of her cancer cells publicly available for research and clinical use. Previous cancer cell lines had been released without the explicit consent of the donors, creating potential legal and ethical impediments to their use in research and drug development. “In the past, there have been controversies about using genetic data for research due to the lack of consent by patients,” said NIST biomedical engineer Justin Zook. “This is the first cancer cell line developed from an individual who explicitly consented to making her genomic data freely available to the public.”
To analyze the genome of pancreatic cancer cells, NIST researchers used more than a dozen distinct state-of-the-art whole genome measurement technologies, some of which were only recently developed. The resulting NIST data on this cancer genome—that is, the full set of genetic instructions from the cell, including the mutations that caused the disease—is several terabytes in size.
The complete genomic data was released by the NIST Genome in a Bottle (GIAB) Consortium, a 12-year-old public-private-academic collaboration to develop reference standards and protocols for genetic analysis and sequencing in research and clinical practice. The NIST scientists suggest the data could be used to research tumors, help to improve cancer diagnostic tests, and potentially aid in the development of new cancer treatments.
The researchers described the cell line and genomic data in Scientific Data (“Development and extensive sequencing of a broadly-consented Genome in a Bottle matched tumor-normal pair”). In their paper, the team stated, “We expect these extensive data sets to be useful for methods development for somatic variant calling, personalized tumor-normal analysis, and tumor genome assembly.”
In 2022, NIST started the Cancer Genome in a Bottle program as part of the consortium to focus on cancer. The Genome in a Bottle Consortium has previously developed sequencing data and benchmarks for normal human cell lines, and is also developing matched tumor-normal samples, which it says represent “the first explicitly consented for public dissemination of genomic data and cell lines.”
Some tumor cell lines have already been characterized as benchmarks by other groups, but data are from legacy cell lines with no consent, or consent before whole genome sequencing was routine, the study authors wrote. For example, in 1951, doctors harvested cervical cancer cells from Baltimore resident Henrietta Lacks without her consent. She died shortly thereafter and was buried in an unmarked grave. But her cells played a major role in the development of the polio vaccine, genetic research, and even COVID-19 vaccines. The Immortal Life of Henrietta Lacks became a best-selling book and an HBO movie starring Oprah Winfrey.
NIH determined in 2013 that researchers could publicly share genomic data from legacy cancer cell lines in the Cancer Cell Line Encyclopedia despite having no or limited consent, because most of these cell lines already had public genomic data available.
However, with the acknowledgement that this could be revised in the future, NIH also “strongly recommended that informed consent processes for prospective research development of human cell lines (for research or commercial purposes) fully describe and consider any risks associated with broad distribution of genomic data derived from those cell lines.” Therefore, the team noted, there is a need for new tumor-normal pairs with explicit consent for public dissemination of genome sequencing data and cell lines, so that they can be developed into enduring reference samples for benchmarking somatic variants.
For their newly reported study analyzing the genome of pancreatic cancer cells, NIST researchers harnessed different state-of-the-art whole genome measurement approaches. The new immortal pancreatic ductal adenocarcinoma (PDAC) cell line and paired normal pancreatic and duodenal tissues are derived from a female of European ancestry, who has given explicit consent for public release of the genomic data. “Data for the tumor-normal matched samples comes from seventeen distinct state-of-the-art whole genome measurement technologies, including high-depth short and long-read bulk whole genome sequencing (WGS), single cell WGS, Hi-C, and karyotyping,” the investigators noted.
Each of the analytical methods identifies the sequence of DNA nucleotides—adenine (A), cytosine (C), guanine (G), and thymine (T)—in an individual’s genome. However, the methods produce slightly varying results and have different strengths and weaknesses.
NIST’s dataset contains separate results for each of the techniques used to sequence the cancer genome. Scientists performing their analysis can compare their data with NIST’s. If there are discrepancies, they can then determine whether their equipment is working properly and remedy the problem if not.
Researchers can also use the genomic data from this cell line to train artificial intelligence models to detect cancer-causing mutations and determine which drugs might work best for treating those cancers. Health care practitioners send cancer patients’ cells to labs for gene sequencing. The results help them better understand the patient’s illness and identify appropriate treatments.
“Labs can use NIST’s database to perform quality control on their equipment,” said Zook. “As a result, the public can have more confidence in the results produced by clinical laboratories that analyze cancer tumors.”
Scientists can also use NIST’s data to advance cancer research. The nucleotide sequences can be interrogated to identify genetic mutations that may offer new insights into how cancer develops and progresses in humans. In summary, the team noted, “We expect these data to facilitate innovation for whole genome measurement technologies, de novo assembly of tumor and normal genomes, and bioinformatic tools to identify small and structural somatic variants.”
Though certain features of NIST’s analysis pertain only to pancreatic cancer—and only pancreatic cancer in this individual—the genomes of all cancers contain sufficiently similar types of mutations that it will be useful for researching other cancers as well, Zook said.
Technology manufacturers may also analyze NIST’s results to identify the strengths and weaknesses of various gene sequencing technologies currently on the market. Zook said this may lead to improvements in existing methods and the development of new sequencing platforms. “This first-of-its-kind broadly consented open-access resource will facilitate further understanding of sequencing methods used for cancer biology,” the investigators stated.
Having sequenced the genome of this pancreatic cell line, Zook and his colleagues will turn their attention to other types of cancer and plan to release data on those genomes as well. The pancreatic cancer patient who donated her cancer cells also donated her noncancerous ones. Zook’s team has published data from these healthy cells as well, so they can be compared with her tumor cells.
“Long term, we would love to see our research lead to new and better cancer diagnostics and treatments,” Zook said. The genomic data from this cell line is freely available on NIST’s Cancer Genome in a Bottle website.
The post NIST Releases Pancreatic Cancer Panacea of Genomic Content appeared first on GEN – Genetic Engineering and Biotechnology News.