Introduction
Breast cancer (BC) is the most common cancer among Egyptian females, accounting for 18.9% of all malignancies and 38.8% of female cancers1,2. It is currently the second most common cause of Egyptian cancer mortality after hepatocellular carcinoma, with an estimated mortality rate of approximately 11% in 20203. Although the incidence of BC in Egypt is lower than that globally reported3, the mortality rate of BC in Egypt is 19.1 deaths per 100,000 compared to 14.9 deaths per 100,000 in the U.S.4. The mean age of BC in Egyptian patients is 10 years younger than in Western countries5. The increasing morbidity and mortality associated with BC in Egypt represents a special challenge to health authorities3. According to the published national registry, approximately 80% of BC cases in Egypt are sporadic, and 18.7% are familial6. According to the American Cancer Society (ACS), about 5% to 10% of BC cases are thought to be hereditary7, where the involvement of genetic factors as the alterations of the highly penetrant BC-causing genes (BRCA1/2) deserves special evaluation8,9. Until recently, the volume of genetic testing among Egyptian women has been disproportionately low due to a complex interplay of social, economic, and cultural factors governing healthcare access.
Since their discovery, thousands of BRCA1/2 pathogenic/likely pathogenic variants PVs/LPVs have been identified10,11. Many have variable pathogenicity potential, conferring various effects on BRCA1/2 protein function12,13. The prevalence of BRCA1/2 PVs/LPVs varies between ethnic groups and geographical areas, leading to several comprehensive studies in different countries to determine the mutational profile of both BRCA1/ 2 genes14,15. A wide variation in their pathogenic potential in relation to other risk factors has been revealed. The landscape of BRCA1/2 PVs/LPVs accordingly became important for better identifying individuals at increased risk of the disease. The prevalence of germline BRCA1/2 (gBRCA1/2) PVs/LPVs in BC was reported earlier in other Middle East and North Africa (MENA) countries; 12.9% in Saudi Arabia, 15.1% in Morocco, 15.5% in Lebanon and 13.9% in Jordan16,17,18,19 and in other studies15,20,21. The presence of gBRCA1 PVs/LPVs is known to increase the risk of triple-negative BC (TNBC), a more aggressive subtype characterized by the absence of ER, PR, and HER2 expression, and thus do not respond to hormonal therapy or anti-HER2 targeted therapy, which are among the most attainable therapeutic options in Egypt22. In contrast, gBRCA2 PVs/LPVs are more commonly associated with estrogen receptor-positive BC23,24,25.
In Egypt, the socioeconomic and genetic variables underlying the biological factors associated with BC represent significant challenges. The landscape of BRCA1/2 disease-causing variants becomes mandatory to identify individuals at increased risk of the disease. The present study is an exploratory study of the BC population for whom BRCA1/2 testing was indicated according to the National Comprehensive Cancer Network (NCCN) guidelines version 1.2023.
The present study aims
- 1-
To assess the mutational landscape of gBRCA1/2 genes in a cohort of 500 female BC patients.
- 2-
To highlight the correlation between BC subtypes and gBRCA1/2 PVs/LPVs in Egyptian patients.
Methods
Relevant guidelines and regulations are carried out in all methods. All experimental protocols were performed according to the institutional ethical procedures and approved by the Institutional Review Board of the National Cancer Institute (IRB-NCI), Cairo University, Egypt (Date: 24thJan 2023/No. 2301-305-052) in accordance with the ethical standards of the Helsinki Declaration26. A written informed consent was obtained from all individual participants included in the study.
Patient selection
Between January 2023 and January 2025, a total of 500 female BC patients with a mean age of 47.29 ± 13.26 years were examined by the Multidisciplinary team of the Oncology Department (MDT) of Cairo University. Immunohistochemistry (IHC) data, including tumor stage, Ki-67 index, histological and molecular subtypes, were obtained from pathology reports issued by the Pathology Department as part of routine diagnostic assessment. HR+ tumors were defined as those expressing ER and/or PR. Triple-negative tumors were negative for ER, PR, and HER2. HER2-positive tumors were excluded from the study. According to IHC data, patients with hormone receptor-positive (HR+) BC constituted 195 cases (39%), while 305 patients (61%) had hormone receptor–negative (HR−) BC. Among the HR− group, 268 patients (53%) had TNBC, and 37 patients had low estrogen receptor (ER) (1–10%) and/ or low progesterone receptor (PR) expression with HER2 negative status. For these patients, gBRCA1/2 testing was indicated according to the guidelines of the NCCN version 1.2023 and the Olympia study. Patients in this study were selected if they fulfilled at least one of the following criteria: early onset BC, defined as BC diagnosis at an age below 40 years27,28, a history of BC and/or other related cancer like ovarian or male BC cancer in one or more patient family members at age below 50 years, or BC with advanced tumor stage or high grade (Stage III-IV based on American Joint Committee on Cancer (AJCC)29. Patients with non-primary BC, incomplete clinical data, poor DNA quality, and those with confirmed HER2 positive phenotype were excluded. IHC data, including tumor stage, Ki-67 index, and histological type, were obtained from pathology reports issued by the Pathology Department as part of routine diagnostic assessment.
DNA extraction and next generation sequencing
According to the manufacturer’s instructions, genomic DNA was extracted from peripheral blood via a QIAamp DNA Blood Mini Kit (Qiagen, Germany). DNA quality was checked and optimized using Qubit® 2.0 Fluorometer (Invitrogen, USA) for Polymerase chain reaction (PCR)-based library preparation with BRCA Pro Panel (Amoy Diagnostics Ltd, Xiamen, Fujian, China) according to manufacturer instructions. Libraries were sequenced on the MiSeq platform (Illumina), enabling the detection of germline SNVs/Indels and large germline rearrangements. This panel comprehensively covers all protein-coding regions, intron/exon boundaries, introns, and untranslated regions (UTRs) of the BRCA1 (NM_007294.4) and BRCA2 (NM_000059.4) genes. The amplified regions of BRCA1/2 were aligned to hg-19 human reference to detect deletions, insertions, and substitutions with all VCF files available. The analytical accuracy of the base calls was > 90%. The minimum coverage was 200× , the uniformity was 95%, and the accepted Q30 was over 85% bases. The variant allele frequency (VAF) of the detected PVs/LPVs was 30–60%. The coding exons and intron/exon junctions of BRCA1/2 were optimally covered and sequenced according to European Molecular Quality Network (EMQN) best practice guidelines for genetic testing30.
Data analysis
All detected variants were interpreted according to the updated American College of Medical Genetics (ACMG) and the Association for Molecular Pathology (AMP) guidelines for BRCA1/2 genes31. Each variant was cross-checked against ClinVar and dbSNP databases, and novel variants were further validated for nomenclature accuracy and pathogenicity using in silico tools VarSome and Franklin. The categories of classification included pathogenic (class 5), likely pathogenic (class 4), variants of uncertain significance (VUS) (class 3), likely benign (class 2), and benign (class 1). Sequencing data were analyzed using the AmoyDx NGS Data Analysis System (ANDAS), a validated and CE–IVD–certified secondary and tertiary bioinformatics analysis pipeline. ANDAS performs automated demultiplexing, quality control, alignment to the human reference genome (GRCh37/hg19), variant calling, and annotation. It is specifically optimized for the AmoyDx BRCA panel to ensure high sensitivity and specificity in detecting BRCA1/2 pathogenic and likely pathogenic variants. Additional in silico prediction software and external curated databases, including BC information core (BIC), ClinVar, ClinGen, and Varsome, were used for better assignment of pathogenicity32,33,34,35,36,37.
Statistical analysis
Statistical analysis was performed using IBM SPSS® Statistics version 22 (IBM® Corp., Armonk, NY, USA). Pearson’s chi-square (χ2) or Fisher’s exact tests were used to compare the clinicopathological data between patients with BRCA1 PVs/LPVs and patients with BRCA2 PVs/LPVs to estimate the association between hormonal status and patient age. A p-value ≤ 0.05 was considered statistically significant.
Results
Demographic and pathological characteristics of the study cohort
500 BC patients with invasive ductal carcinoma (IDC) as a prevalent histotype (88.8%) were enrolled in this study. Their mean age was 47.29 ± 13.26 years, and the median was 44 years (19–84). The clinical and pathological features of the whole cohort are summarized in Table 1.
There was a significant difference in age at diagnosis between BRCA1/2 PV/LPV carriers and non-carriers (42.9 ± 11.6 vs. 48.0 ± 13.4 years; p = 0.0028). A significant age difference was also observed between BRCA1 and BRCA2 PV/LPV carriers, with BRCA1 carriers being younger than BRCA2 carriers (39.1 ± 8.71 vs. 48.25 ± 12.79 years; p = 0.004). Germline BRCA1/2 PVs/LPVs were identified in 58 out of 500 BC patients (11.6%). Of these, 34 patients (6.8%) carried gBRCA1 PVs/LPVs, and 24 (4.8%) carried gBRCA2 PVs/LPVs. A history of BC and/or other related cancer in one or more patient family members at an age below 50 years was reported in 352 patients (70.4%). Among 268 patients with TNBC, 41 patients (15.3%) were carriers of gBRCA1/2 PVs/LPVs, of whom 28 patients (10.4%) with gBRCA1 PVs/LPVs and 13 patients (4.9%) with gBRCA2 PVs/LPVs (Tables 1 and 2). Patients with TNBC had higher BRCA1/2 PVs/LPVs rate (8.2%) than those with HR+ BC (3.4%). This patient category had the highest rates of proliferating tumor index (Ki67>20), family history of BC or related cancer, and earliest onset disease (age younger than 40).
Distribution of BRCA1/2 variants
Based on the standards and guidelines for interpreting sequence variants31, we identified 55 BRCA1/2 PVs/LPVs (including 44 SNVs in addition to 11 CNVs), of which 44 were reported in the Clinvar database37, and 11 were considered novel variants.
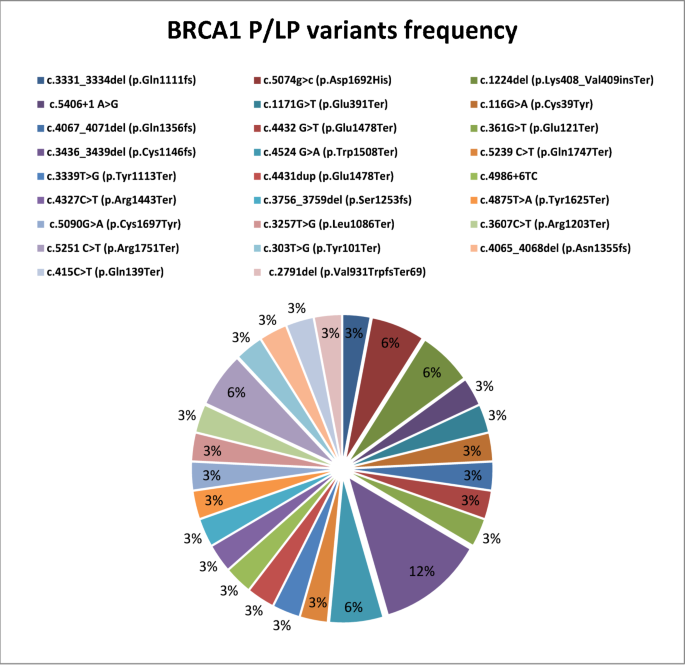
Twenty-seven P/LP BRCA1 SNVs were detected, including 13 nonsense, 7 frameshifts, 5 missense, and 2 intronic variants (Fig. 1). Additionally, 17 BRCA2 P/LP SNVs were identified, including 11 frameshift, 3 nonsense, 2 missense, and 1 intronic variant (Fig. 2). Nonsense variants were prevalent in BRCA1 (13/27, 48.1%), whereas frameshift variants were more prevalent in BRCA2 (11/17, 64.7%). Patients carriers of gBRCA1 PVs/LPVs were characterized by younger age (< 40 years, 61.8%) and positive family history (85.3%).
Three novel LPVs gBRCA1 were identified, namely, c.361G>T (p.Glu121Ter) (patient A75), c.2791del (p.Val931TrpfsTer69) (patient M75) and c.4431dup (p.Glu1478Ter) (patient M338) located in exons 6, 10 and 11 respectively.
In BRCA2 gene, two novel frameshift LPVs were identified in exon 11; c.3139del (Ile1047LeufsTer2) (patient H418) and c.5690dup (p.Leu1897PhefsTer3) (patient M476) (Tables 3 and 4).
Copy number variations (CNVs)
Software-based copy number analysis identified 12 variants in the BRCA1/2 genes, of which 5 were previously reported in the literature15,37,38,39,40,41,42 (Table 5).
Five novel BRCA1 CNVs involving losses or gains of crucial parts of the gene were detected, including loss of exons 2–20, gain of exon 6, gain of exons 14–16, gain of exon 21, and duplication of exons 20–24. Additionally, two novel BRCA2 CNVs were detected; a frameshift variant due to exon 11 deletion and an in-frame insertion resulting from exon 12 gain. Both variants contribute to genomic instability, which may have implications for cancer development and progression.
Variants of uncertain significance (VUS)
Nineteen VUS were recorded in 22 (4.4%) patients; 8 gBRCA1 and 14 gBRCA2 VUS (Table 6). Thirteen patients (59%) with VUS had a positive family history of BC.
Discussion
The present cohort represents a referral population from different Egyptian governorates, reflecting diverse personal and familial risk factors. The overall prevalence of gBRCA1/2 PVs/LPVs of 11.6% is not far from that reported in other MENA countries16,17,18,19. However, in China, a wide variation in the prevalence of gBRCA1/2 PVs/LPVs has been reported among patients with high‐risk BC, ranging from 8.3% to 17.6%43,44.
In our cohort, 8.2% of TNBC patients were carriers of gBRCA1/2 PVs/LPVs, whereas 9.3% were reported in Australia and 15.4% in the United States45. Consistent with other studies, gBRCA1 PVs/LPVs were more frequently detected in patients with TNBC- whose tumors are unresponsive to hormonal or HER2-targeted therapies—than gBRCA2 PVs/LPVs, representing 10.4% vs. 5.2%, respectively22,46,47. The patient category with gBRCA1 PVs/LPVs was characterized by younger age (< 40 years, 61.8%) and positive family history (85.3%), findings that should draw special attention to the need for genetic counseling programs.
The two reported novel gBRCA1 LPVs are located in exon 11 and are expected to confer a nonsense-mediated decay of RNA predisposing to differential breast or ovarian cancer risks and early onset BC48. Two patients had a strong family history of BC and the third developed contralateral BC.
Four patients (S204, H219, S271, T315) were carriers of the same pathogenic gBRCA1 variant, c.3436_3439del (p.Cys1146fs), previously described in Slovenia, China, and Lebanon49,50,51. These patients were originating from different unrelated families. However, it is common for recurrent mutations to be identified at elevated frequencies across different ethnic groups52.
Most pathogenic gBRCA2 variants were located in exon 11 and expected to disrupt the interaction of BRCA2 with other proteins, such as RAD51 and TP53, which work together to mediate DNA damage repair53. Seventeen BRCA2 PVs/LPVs were previously reported in universal databases54. The BRCA2 novel mutations, c.3139del (Ile1047LeufsTer2) and c.5690dup (p.Leu1897PhefsTer3), are located within the BCCR1 and BCCR2 regions, respectively. These regions are thought to mediate interactions with key transcription factors and DNA repair proteins, potentially disrupting crucial transcription factors and DNA repair proteins55,56. Notably, several of the BRCA1/2 PVs/ LPVs identified in our cohort have also been reported in other studies from the MENA region15,17,18,19,20,21.
In the literature, most large genomic rearrangements (LGRs) are pathogenic and are more commonly found within the BRCA1 gene than in BRCA257. In our study, variants with abnormal copy numbers (CNVs) represented 18.6% of all variants, with more BRCA1 CNVs (12.9%) than BRCA2 CNVs (5.7%). A Dutch study reported 27% BRCA1 CNVs of all gBRCA1 variants58. Another study in China reported an incidence of 16.1% in BRCA1 CNVs59. BRCA1 CNVs involve gains or losses of crucial parts of the gene involving domains expected to affect genomic instability as the BRCT and c-terminal domains. They are associated with elevated BC risk, especially for patients with BRCA1 deletions60. Screening CNVs in BC patients who tested negative for gBRCA1/2 variants may indicate different therapeutic options, such as targeted therapy or immunotherapy.
The present study reported 22 (4.4%) patients with BRCA1 and BRCA2 VUS. The rate of VUS varies from 3.3% to 4.4% across different races and is the highest in the Asian population, with up to 15%36,61,62,63. The presence of VUS in 22 of our patients can be hard to ignore, primarily if a compelling personal and/or family history exists. Regularly updating variant databases within this context may help better define the clinical significance of VUS reclassification.
In the present study, 70% of patients had a positive family history of breast/ovarian cancer. In contrast, carriers of gBRCA1/2 PVs/LPVs represented only 11.6% of the BC cohort, which points to the stronger influential role of family history as BC predisposing64,65 and highlights the need to explore other genetic factors in patients who test negative for BRCA1/2 PVs/LPVs. Consistent with other studies, applying multigene panel testing of other BC predisposing genes with variable penetrance, such as PALB2, PTEN, ATM, STK11, and CHEK2, may provide a better comprehensive genomic picture of BC66.
The fact that approximately 26,000 new BC cases per year in Egypt are expected to reach 46,000 cases by 20503 represents a challenge to health authorities regarding the underlying causes. It implies a need for more comprehensive BC screening programs that pinpoint early-onset BC cases. The routine application of genetic studies and implementation of counseling programs for this patient category is recommended.
Limitations
This study has some limitations. Despite being one of the most extensive BRCA1/2 genetic screening studies among Egyptian BC patients, the sample size of 500 remains relatively small compared to Egypt’s large population of over 100 million. As a result, the sample size may not fully capture the prevalence of rare variants or represent the full genetic diversity of the broader Egyptian population.
Conclusion
Our study ranks among the few studies in Egypt that explore the prevalence and spectrum of germline BRCA1/2 gene variants. Our findings point to the need for extended multigene profiling as part of routine BC risk assessment.
Data availability
VCF files are available from the corresponding author upon reasonable request. The data of current study are involved in the article.The patients’ clinicopathological data are presented in Tables 1 and 2, the detected SNPs and their accession numbers (rs) are illustrated in Tables 3, 4, 6) in the manuscript. The datasets and accession numbers generated and/or analysed during the current study are available in the [dbSNP] repository (https://www.ncbi.nlm.nih.gov/snp/) and [ClinVar] repository (https://www.ncbi.nlm.nih.gov/clinvar/)”.
Abbreviations
- ACS:
-
American cancer society
- BC:
-
Breast cancer
- NCCN:
-
National comprehensive cancer network
- AJCC:
-
American joint committee on cancer
- ACMG:
-
American college of medical genetics
- HR:
-
Hormone receptor
- TNBC:
-
Triple negative breast cancer
- P/LP:
-
Pathogenic/likely pathogenic
- PVs/LPVs:
-
Pathogenic variants/likely pathogenic variants
- g BRCA :
-
Germilne BRCA
- SNVs:
-
Single nucleotide variants
- CNVs:
-
Copy number variations
- HGVS:
-
Human genome variation society
- AMP:
-
Association for molecular pathology
- BIC:
-
Breast cancer information core
- IDC:
-
Invasive ductal carcinoma
- MENA:
-
Middle East and North Africa
- BCCR:
-
Breast cancer cluster region
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor 2
- LRGs:
-
Large genomic rearrangements
- EMQN:
-
European molecular quality network
- IRB:
-
Institutional review board
References
-
Mahfouz, E. M. Clinical characteristics of breast cancer in young women ≤ 40 years old-Minia-Egypt. Minia J. Med. Res. 31(4), 221–227 (2020).
-
Alorabi, M., Elghazawy, H. et al. Cancer control in Egypt: Investing in health. (The ASCO Post, 2021).
-
Azim, H. A. et al. Clinicopathologic features of breast cancer in Egypt—contemporary profile and future needs: A systematic review and meta-analysis. JCO Glob. Oncol. 9, e2200387 (2023).
-
Ahmed, H. F. et al. Cost effectiveness analysis of breast cancer screening in upper-and low-middle income countries focusing on Egypt: A systematic review. J. Pharm. Negative Results https://doi.org/10.47750/pnr.2023.14.02.24 (2023).
-
Mohammed, M. A. et al. Retrospective descriptive analysis of the demographic and clinicopathological presentation of breast cancer patients in Kasr Al-Ainy Hospital over 5 years. Egypt. J. Surg. 43(2), 515–523 (2024).
-
Farahat, T. M. et al. Effect of family history on clinical and pathological characteristics of breast cancer: A hospital-based study in Egypt. Egypt. Family Med. J. 1(2), 1–14 (2017).
-
American Cancer Society. (n.d.). Breast cancer risk factors you cannot change. Retrieved from https://www.cancer.org/cancer/breast-cancer/risk-and-prevention/breast-cancer-risk-factors-you-cannot-change.html
-
Petrucelli, N., Daly, M. B. et al. BRCA1-and BRCA2-associated hereditary breast and ovarian cancer (2022).
-
Wu, S. et al. Molecular mechanisms of PALB2 function and its role in breast cancer management. Front. Oncol. 10, 301 (2020).
-
Cline, M. S. et al. BRCA Challenge: BRCA exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 14(12), e1007752 (2018).
-
Li, J. et al. Human BRCA pathogenic variants were originated during recent human history. Life Sci. Alliance 5(5), e202101263 (2022).
-
Mahdavi, M. et al. Hereditary breast cancer; Genetic penetrance and current status with BRCA. J. Cell. Physiol. 234(5), 5741–5750 (2019).
-
Venkitaraman, A. R. How do mutations affecting the breast cancer genes BRCA1 and BRCA2 cause cancer susceptibility?. DNA Repair 81, 102668 (2019).
-
Herzog, J. S. et al. Genetic epidemiology of BRCA1-and BRCA2-associated cancer across Latin America. NPJ Breast Cancer 7(1), 107 (2021).
-
ElBiad, O. et al. Prevalence of specific and recurrent/founder pathogenic variants in BRCA genes in breast and ovarian cancer in North Africa. BMC Cancer 22(1), 208 (2022).
-
Abulkhair, O. et al. Prevalence of BRCA1 and BRCA2 mutations among high-risk Saudi patients with breast cancer. J. Global Oncol. 4, 1–9 (2018).
-
Bakkach, J. et al. Contribution of BRCA1 and BRCA2 germline mutations to early onset breast cancer: A series from north of Morocco. BMC Cancer 20, 1–8 (2020).
-
Jalkh, N. et al. Next-generation sequencing in familial breast cancer patients from Lebanon. BMC Med. Genom. 10, 1–12 (2017).
-
Abdel-Razeq, H. et al. Patterns and prevalence of germline BRCA1 and BRCA2 mutations among high-risk breast cancer patients in Jordan: A study of 500 patients. J. Oncol. 2020(1), 8362179 (2020).
-
Laitman, Y. et al. The spectrum of BRCA1 and BRCA2 pathogenic sequence variants in Middle Eastern, North African, and South European countries. Hum. Mutat. 40(11), e1–e23 (2019).
-
Azadnajafabad, S. et al. Burden of breast cancer and attributable risk factors in the North Africa and Middle East region, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Front. Oncol. 13, 1132816 (2023).
-
Kim, J. et al. Mutations of TP53 and genes related to homologous recombination repair in breast cancer with germline BRCA1/2 mutations. Hum. Genom. 17(1), 2 (2023).
-
Lee, E. et al. Characteristics of triple-negative breast cancer in patients with a BRCA1 mutation: Results from a population-based study of young women. J. Clin. Oncol. 29(33), 4373–4380 (2011).
-
Glodzik, D. et al. Comprehensive molecular comparison of BRCA1 hypermethylated and BRCA1 mutated triple negative breast cancers. Nat. Commun. 11(1), 3747 (2020).
-
Vidarsdottir, L. et al. Estrogen receptor-positive breast cancer and adverse outcome in BRCA2 mutation carriers and young non-carrier patients. NPJ Breast Cancer 9(1), 95 (2023).
-
Ashcroft, R. E. The declaration of Helsinki. In The Oxford textbook of clinical research ethics 141–148 (Oxford University Press, New York, 2008).
-
Brenner, D. R. et al. Projected estimates of cancer in Canada in 2022. CMAJ 194(17), E601–E607 (2022).
-
Basmadjian, R. B. et al. The association between early-onset diagnosis and clinical outcomes in triple-negative breast cancer: A systematic review and meta-analysis. Cancers 15(7), 1923 (2023).
-
Edge, S. B. & Compton, C. C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 17(6), 1471–1474 (2010).
-
McDevitt, T. et al. EMQN best practice guidelines for genetic testing in hereditary breast and ovarian cancer. Eur. J. Hum. Genet. 32(5), 479–488 (2024).
-
Parsons, M. T. et al. Evidence-based recommendations for gene-specific ACMG/AMP variant classification from the ClinGen ENIGMA BRCA1 and BRCA2 variant curation expert panel. Am. J. Human Genet. 111(9), 2044–2058 (2024).
-
Møller, P. et al. A simplified method for segregation analysis (SISA) to determine penetrance and expression of a genetic variant in a family. Hum. Mutat. 32(5), 568–571 (2011).
-
Consortium, G. P. A global reference for human genetic variation. Nature 526(7571), 68 (2015).
-
McLaren, W. The ensembl variant effect predictor. Genom. Biol. 17, 122 (2016).
-
Gunning, A. C. et al. Assessing performance of pathogenicity predictors using clinically relevant variant datasets. J. Med. Genet. 58(8), 547–555 (2021).
-
Kwong, A. et al. How does re-classification of variants of unknown significance (VUS) impact the management of patients at risk for hereditary breast cancer?. BMC Med. Genom. 15(1), 122 (2022).
-
Landrum, M. J. et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46(D1), D1062–D1067 (2018).
-
Del Valle, J. et al. Identification and comprehensive characterization of large genomic rearrangements in the BRCA1 and BRCA2 genes. Breast Cancer Res. Treatment 122, 733–743 (2010).
-
Fachal, L. et al. Large genomic rearrangements of BRCA1 and BRCA2 among patients referred for genetic analysis in Galicia (NW Spain): Delimitation and mechanism of three novel BRCA1 rearrangements. PLoS ONE 9(3), e93306 (2014).
-
Hua, D. et al. Next-generation sequencing based detection of BRCA1 and BRCA2 large genomic rearrangements in Chinese cancer patients. Front. Oncol. 12, 898916 (2022).
-
Ea, V. et al. BRCA1 intragenic duplication combined with a likely pathogenic TP53 variant in a patient with triple-negative breast cancer: Clinical risk and management. Int. J. Mol. Sci. 25(11), 6274 (2024).
-
Béroud, C. et al. BRCA share: A collection of clinical BRCA gene variants. Hum. Mutat. 37(12), 1318–1328 (2016).
-
Wang, X. et al. Prevalence of BRCA1 and BRCA2 gene mutations in Chinese patients with high-risk breast cancer. Mol. Genet. Genomic Med. 7(6), e677 (2019).
-
Zhang, Y. et al. BRCA1 and BRCA2 germline mutations in Chinese Hakka breast cancer patients. BMC Med. Genomics 17(1), 3 (2024).
-
Armstrong, N. et al. A systematic review of the international prevalence of BRCA mutation in breast cancer. Clin. Epidemiol. 11, 543–561 (2019).
-
Park, S. et al. Clinical characteristics and exploratory genomic analyses of germline BRCA1 or BRCA2 mutations in breast cancer. Mol. Cancer Res. 18(9), 1315–1325 (2020).
-
Yin, L. et al. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 22, 1–13 (2020).
-
Rebbeck, T. R. et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA 313(13), 1347–1361 (2015).
-
Novaković, S. et al. Novel BRCA1 and BRCA2 pathogenic mutations in Slovene hereditary breast and ovarian cancer families. Int. J. Oncol. 41(5), 1619–1627 (2012).
-
Jian, W. et al. Clinical and genetic characterization of hereditary breast cancer in a Chinese population. Hered. Cancer Clin. Pract. 15, 1–9 (2017).
-
Farra, C. et al. BRCA mutation screening and patterns among high-risk Lebanese subjects. Hered. Cancer Clin. Pract. 17, 1–7 (2019).
-
Rebbeck, T. R. et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 39(5), 593–620 (2018).
-
Shailani, A. et al. A comprehensive analysis of BRCA2 gene: Focus on mechanistic aspects of its functions, spectrum of deleterious mutations, and therapeutic strategies targeting BRCA2-deficient tumors. Med. Oncol. 35(3), 18 (2018).
-
Johnston, J. J. et al. Secondary variants in individuals undergoing exome sequencing: Screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am. J. Human Genet. 91(1), 97–108 (2012).
-
Clark, S. L. et al. Structure-function of the tumor suppressor BRCA1. Comput. Struct. Biotechnol. J. 1(1), e201204005 (2012).
-
Elshwekh, H. et al. Assessing the impact of novel BRCA1 exon 11 variants on pre-mRNA splicing. Cells 13(10), 824 (2024).
-
Caputo, S. M. et al. 5′ region large genomic rearrangements in the BRCA1 Gene in French families: Identification of a Tandem triplication and nine distinct deletions with five recurrent breakpoints. Cancers 13(13), 3171 (2021).
-
Schmidt, A. Y. et al. Next-generation sequencing-based detection of germline copy number variations in BRCA1/BRCA2: Validation of a one-step diagnostic workflow. J. Mol. Diagn. 19(6), 809–816 (2017).
-
El Ansari, F. Z. et al. Screening of BRCA1/2 genes mutations and copy number variations in patients with high risk for hereditary breast and ovarian cancer syndrome (HBOC). BMC Cancer 20, 1–11 (2020).
-
Ismail, T. et al. BRCA1 and its vulnerable C-terminal BRCT domain: Structure, function, genetic mutations and links to Diagnosis and treatment of breast and ovarian cancer. Pharmaceuticals 17(3), 333 (2024).
-
Frank, T. S. et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: Analysis of 10,000 individuals. J. Clin. Oncol. 20(6), 1480–1490 (2002).
-
Caswell-Jin, J. L. et al. Racial ethnic differences in multiple-gene sequencing results for hereditary cancer risk. Genet. Med. 20(2), 234–239 (2018).
-
Lindor, N. M. et al. BRCA1/2 sequence variants of uncertain significance: A primer for providers to assist in discussions and in medical management. Oncologist 18(5), 518–524 (2013).
-
Gabaldó Barrios, X. et al. Molecular characterization and clinical interpretation of BRCA1/BRCA2 variants in families from Murcia (south-eastern Spain) with hereditary breast and ovarian cancer: Clinical–pathological features in BRCA carriers and non-carriers. Fam. Cancer 16, 477–489 (2017).
-
Rweyemamu, L. P. et al. Breast cancer in East Africa: Prevalence and spectrum of germline SNV/indel and CNVs in BRCA1 and BRCA2 genes among breast cancer patients in Tanzania. Cancer Med. 12(3), 3395–3409 (2023).
-
Bono, M. et al. Impact of deleterious variants in other genes beyond BRCA1/2 detected in breast/ovarian and pancreatic cancer patients by NGS-based multi-gene panel testing: Looking over the hedge. ESMO Open 6(4), 100235 (2021).
Acknowledgements
Not applicable.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Cairo university National cancer institute IRB approval number: 2301–305-051.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Elmetnawy, W., Nader, H., ElNahas, T. et al. Mutational spectrum of BRCA genes in Egyptian patients with breast cancer. Sci Rep 15, 26067 (2025). https://doi.org/10.1038/s41598-025-09810-5
-
Received:
-
Accepted:
-
Published:
-
DOI: https://doi.org/10.1038/s41598-025-09810-5