- Research Briefing
- Published:
Subjects
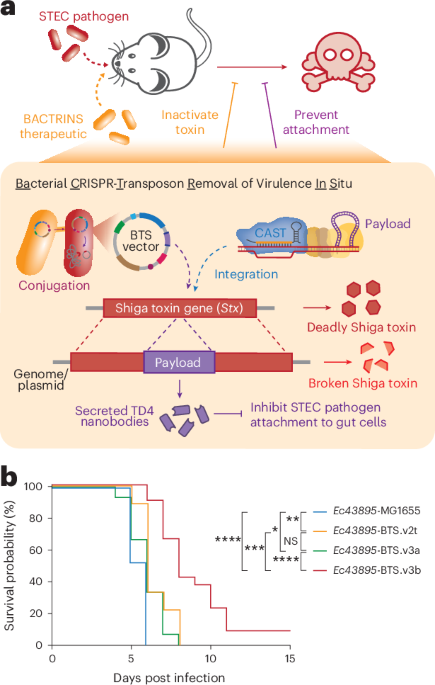
Treatment of severe gastrointestinal illness caused by Shiga-toxin-producing enteropathogenic bacteria is a growing challenge owing to increasing antimicrobial resistance. We engineered a conjugative CRISPR-associated transposase to insert a genetic payload that inactivates the Shiga toxin gene of Enterobacteriaceae pathogens in the gut, establishing a foundation for microbial gene therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

References
-
Gencay, Y. E. et al. Engineered phage with antibacterial CRISPR–Cas selectively reduce E. coli burden in mice. Nat. Biotechnol. 42, 265–274 (2024). This study reports the development and evaluation of engineered phages armed with CRISPR–Cas systems that selectively reduce pathogenic E. coli.
-
Ronda, C., Chen, S. P., Cabral, V., Yaung, S. J. & Wang, H. H. Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods 16, 167–170 (2019). This paper describes the initial demonstration of microbiota engineering directly in the gut.
-
Vo, P. L. H. et al. CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat. Biotechnol. 39, 480–489 (2021). This paper describes RNA-guided transposases that allow high-efficiency and multiplexed genome editing in bacteria.
-
Ruano-Gallego, D. et al. A nanobody targeting the translocated intimin receptor inhibits the attachment of enterohemorrhagic E. coli to human colonic mucosa. PLoS Pathog. 15, e1008031 (2019). This study identifies and characterizes a nanobody capable of inhibiting attachment of enterohaemorrhagic E. coli to human intestinal tissues.
-
Lentsch, V. et al. Vaccine-enhanced competition permits rational bacterial strain replacement in the gut. Science 388, 74–81 (2025). This study used oral vaccination with engineered competitor E. coli strains to trigger immune-mediated selective replacement of pathogenic E. coli in the gut.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is a summary of: Ronda, C. et al. Precise virulence inactivation using a CRISPR-associated transposase for combating Enterobacteriaceae gut pathogens. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-025-01453-1 (2025).
Rights and permissions
About this article
Cite this article
A live, programmable microbiota therapeutic disables virulence in gut pathogens. Nat. Biomed. Eng (2025). https://doi.org/10.1038/s41551-025-01452-2
-
Published:
-
DOI: https://doi.org/10.1038/s41551-025-01452-2