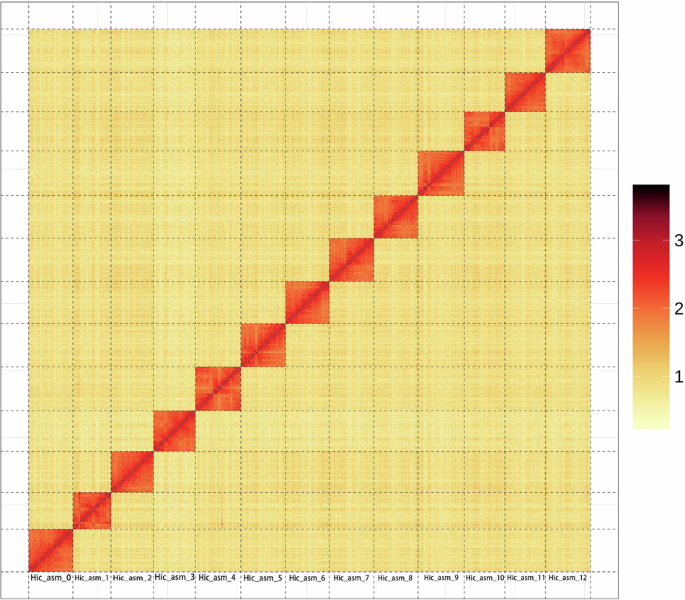
Code availability
All software and pipelines utilized in this study adhered rigorously to the manuals and protocols of the established bioinformatic tools. The specific versions of the software are detailed in the Methods section. If parameters were not specified, default settings were used. No custom code was used in this study.
Data availability
References
-
United Nations Department of Economic and Social Affairs. Population Division (Ed.). World Population 9, 1–54 (2022).
-
Williamson, E., Ross, I. L., Wall, B. T. & Hankamer, B. Microalgae: Potential novel protein for sustainable human nutrition. Trends Plant Sci. 29, 370–382 (2024).
-
Torres-Tiji, Y., Fields, F. J. & Mayfield, S. P. Microalgae as a future food source. Biotechnol Adv. 41, 107536 (2020).
-
Becker, E. W. Micro-algae as a source of protein. Biotechnol Adv. 25, 207–210 (2007).
-
Krienitz, L., Huss, V. A. & Bock, C. Chlorella: 125 years of the green survivalist. Trends Plant Sci. 20, 67–69 (2015).
-
Barbosa, M. J., Janssen, M., Sudfeld, C., D’Adamo, S. & Wijffels, R. H. Hypes, hopes, and the way forward for microalgal biotechnology. Trends Biotechnol. 41, 452–471 (2023).
-
Fan, J. et al. Genomic foundation of starch-to-lipid switch in oleaginous Chlorella spp. Plant Physiol. 169, 2444–2461 (2015).
-
Zou, S. et al. Combining and comparing coalescent, distance and character-based approaches for barcoding microalgaes: A test with Chlorella-like species (Chlorophyta). PLoS One 11, e0153833 (2016).
-
Zhu, T. et al. Transcriptomic and metabolomic analysis reveal the effects of light quality on the growth and lipid biosynthesis in Chlorella pyrenoidosa. Biomolecules. 14, 1144 (2024).
-
Chen, L., Zhang, L. & Liu, T. Concurrent production of carotenoids and lipid by a filamentous microalga Trentepohlia arborum. Bioresource Technol. 214, 567–573 (2016).
-
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 18, 170–175 (2021).
-
Do, V. H. et al. Pasa: leveraging population pangenome graph to scaffold prokaryote genome assemblies. Nucleic Acids Res. 52, e15 (2024).
-
Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Current protocols in bioinformatics Chapter 4, Unit 4.10 (2004).
-
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
-
Chan, P. P., Lin, B. Y., Mak, A. J. & Lowe, T. M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49, 9077–9096 (2021).
-
Kalvari, I. et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 49, D192–D200 (2021).
-
Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019).
-
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
-
Saier, M. H. et al. The Transporter Classification Database (TCDB): 2021 update. Nucleic Acids Res. 49, D461–D467 (2021).
-
Boeckmann, B. et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–70 (2003).
-
Blin, K. et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51, W46–W50 (2023).
-
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP613830 (2025).
-
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP637498 (2025).
-
Zhang, S. et al. The GSA Family in 2025: A broadened sharing platform for multi-omics and multimodal data. Genom Proteom Bioinf. 23, qzaf072 (2025).
-
CNCB-NGDC Members and Partners. Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2025. Nucleic Acids Res. 53, D30–D44 (2025).
-
CNCB Genome Sequence Archive https://ngdc.cncb.ac.cn/gsa/browse/CRA033078 (2025).
-
Luo, X. M. GenBank https://identifiers.org/ncbi/insdc.gca:GCA_047663505.1 (2025).
-
Luo, X. M. Auxenochlorella pyrenoidosa genome annotation and protein sequences. figshare https://doi.org/10.6084/m9.figshare.28498355 (2025).
Acknowledgements
This work was supported by the National Key R and D Program of China (2025YFE0199700, 2023YFE0199400), Agricultural Science and Technology Innovation Program (ASTIP) (No. Y2024QC33), Chengdu Science and Technology Program (No. 2024-YF06-00116-HZ, 2024-YF06-00124-HZ), the Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences (No. 34-IUA-02).
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, X., Su, H., Guan, G. et al. Chromosome-level genome assembly for an edible protein microalgae Auxenochlorella pyrenoidosa. Sci Data (2025). https://doi.org/10.1038/s41597-025-06358-x
-
Received:
-
Accepted:
-
Published:
-
DOI: https://doi.org/10.1038/s41597-025-06358-x