- Research Briefing
- Published:
Subjects
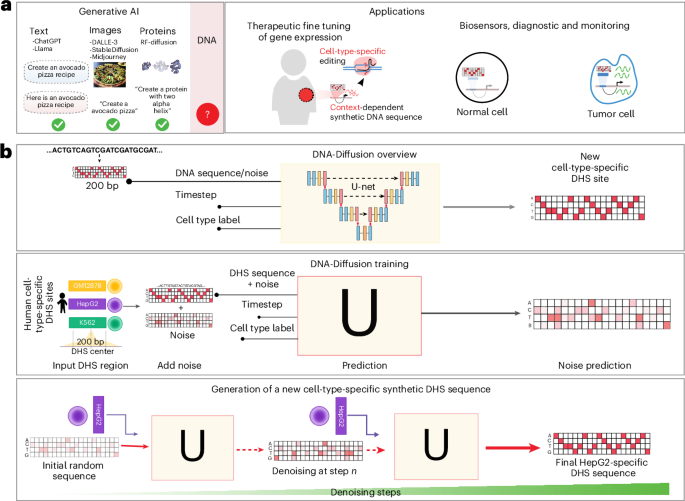
We developed DNA-Diffusion, a generative artificial intelligence (AI) method that creates synthetic regulatory elements showing enhanced activity. Multiple synthetic elements demonstrated superior cell-type-specific expression in computational predictions and episomal assays, and when integrated at AXIN2, a leukemia-protective gene, outperformed naturally occurring protective variants, opening new possibilities for precision gene therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout

References
-
Llimos, G. et al. A leukemia-protective germline variant mediates chromatin module formation via transcription factor nucleation. Nat. Commun. 13, 2042 (2022). This study discovered the protective AXIN2 variant that reduces chronic lymphocytic leukemia progression.
-
Taskiran, I. I. et al. Cell-type-directed design of synthetic enhancers. Nature 626, 212–220 (2024). This paper pioneered generative adversarial network (GAN)-based generative design of cell-type-specific enhancers, establishing the foundation for distribution-learning approaches that DNA-Diffusion builds upon.
-
Gosai, S. J. et al. Machine-guided design of cell-type-targeting cis-regulatory elements. Nature 634, 1211–1220 (2024). This study demonstrated convolutional neural network (CNN)-based optimization for regulatory element design, achieving high signal intensity through direct optimization, which DNA-Diffusion complements by maintaining motif diversity without requiring prior transcription factor knowledge.
-
Ho, J., Jain, A. & Abbeel, P. Denoising diffusion probabilistic models. In Proc. 34th International Conference on Neural Information Processing Systems (eds Larochelle, H. et al.) 6840–6851 (2020). This seminal paper introduced the diffusion model framework that we adapted from image generation to DNA sequences.
-
Kribelbauer-Swietek, J. F. et al. EXTRA-seq: a genome-integrated extended massively parallel reporter assay to quantify enhancer-promoter communication. Preprint at bioRxiv https://doi.org/10.1101/2024.12.08.627402 (2024). This paper presents EXTRA-seq technology, a method that measures how regulatory DNA sequences control gene expression by integrating them into chromosomes and tracking their ability to activate target genes.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is a summary of: DaSilva, L. F. et al. Designing synthetic regulatory elements using the generative AI framework DNA-Diffusion. Nat. Genet. https://doi.org/10.1038/s41588-025-02441-6 (2025).
Rights and permissions
About this article
Cite this article
Generative AI creates synthetic regulatory DNA sequences for precision gene control. Nat Genet (2025). https://doi.org/10.1038/s41588-025-02443-4
-
Published:
-
Version of record:
-
DOI: https://doi.org/10.1038/s41588-025-02443-4