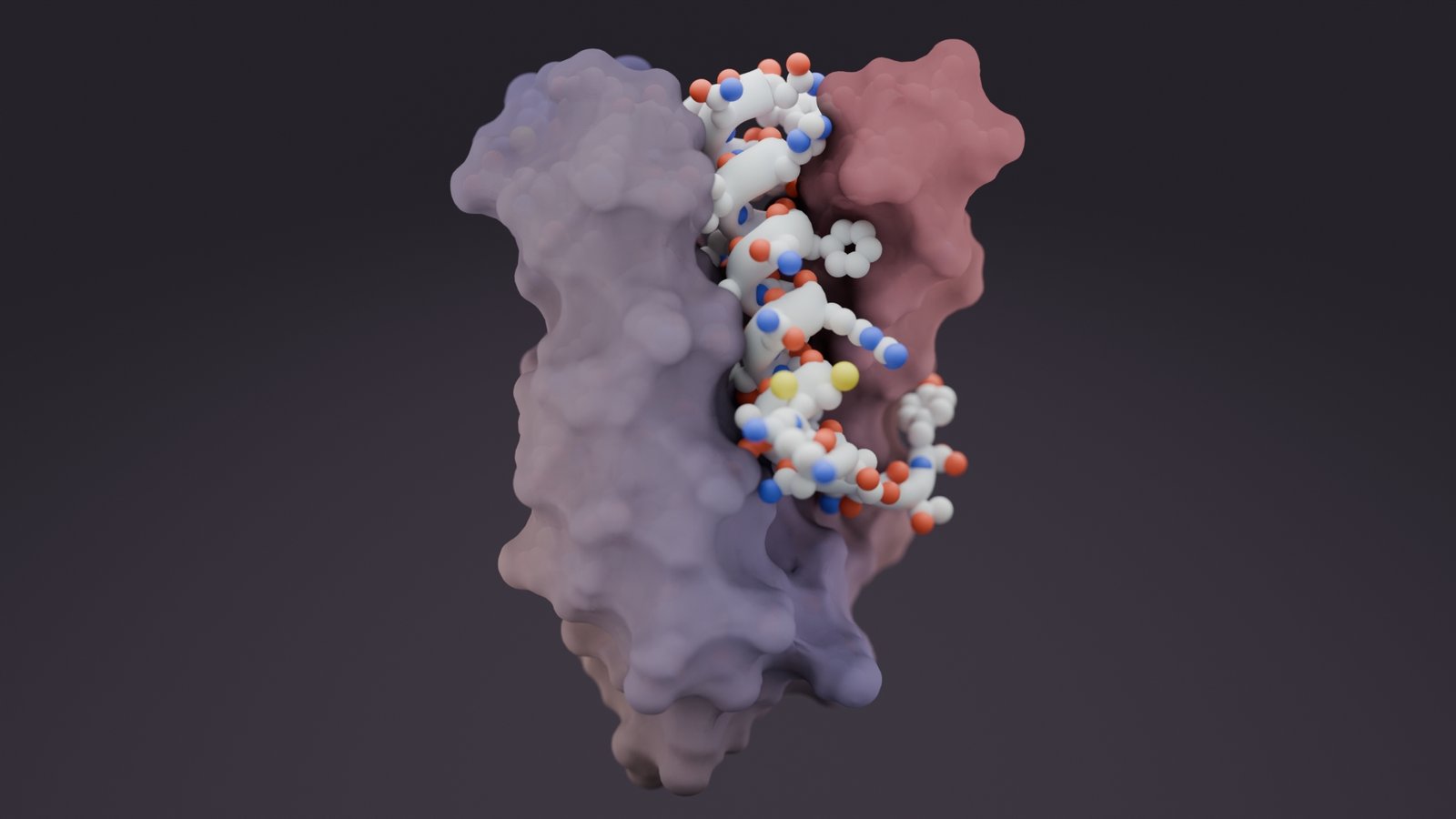
Drug developers have historically glued their attention to targeting the precise and fine-tuned protein structures underlying biological function. Yet this approach misses over 50% of the human proteome, which is highly flexible, flopping between conformations to drive a wealth of disease-related pathways, including pain signaling, cancer progression, and neurological disease. These intrinsically disordered regions (IDRs) have long been untamed by candidate drug binders, flaunting the title, “undruggable”… until now.
Researchers from the lab of Nobel Laureate David Baker, PhD, have now developed complementary AI approaches that design protein binders to IDRs with newfound success rates and affinity. The methods take an amino acid sequence-based approach, which does not require structural information, to facilitate highly generalizable drug discovery on a new trove of therapy targets. The work is documented in a paper published in Science titled “Design of intrinsically disordered region binding proteins,” and a preprint on bioRxiv that has not yet been peer reviewed.
“There’s been a growing appreciation of the importance of disordered proteins in biology, but it’s been very hard to design binders to them,” said Baker in an interview with GEN. “I call this ‘drugging the undruggable.’ This was a very large class of difficult-to-drug targets that we previously could not hit.”
Among the diverse array of IDR-containing proteins that were successfully targeted by the Baker lab included dynorphin, an opioid receptor implicated in chronic pain, a variant of BRCA1, a DNA-repair protein implicated in breast cancer, and amylin, a hormone whose designed binders were shown to dissolve amyloid fibrils associated with type 2 diabetes.
Baker, who is a Howard Hughes Medical Institute (HHMI) investigator and the director of the Institute for Protein Design at the University of Washington (UW), was awarded the 2024 Nobel Prize in Chemistry for his work in pioneering AI for protein design. In recent years, his lab has developed RFdiffusion, a diffusion-based AI model that generates proteins from scratch (de novo) to enable broad applications across cancer immunotherapy, enzyme catalysis, neutralizing snake toxin, and now targeting IDRs.
Easier than ordered proteins
While conventional approaches to target IDRs have tried to develop antibody binders via animal immunizations, IDRs are highly subject to cleavage and aggregation once injected into the animal. Additionally, prior computational methods for binder design relied on pre-specification of the target IDR geometry, thereby limiting generalizability.
To address these challenges, the preprint describes an RFdiffusion-centered approach, which excels at identifying helical and strand secondary structures in IDRs for binder design. This secondary structure propensity is explained by the model’s training on known proteins in the Protein Data Bank (PDB). The generated IDR binders were able to achieve strong nanomolar to picomolar affinities, “matching nature’s strongest interactions,” highlighted Caixuan Liu, PhD, a postdoctoral researcher in the Baker lab and co-lead author of the preprint.
In contrast, the Science paper’s method, termed “logos,” takes a side-chain-centered approach, which creates a library of “pockets” that recognize sets of amino acid side chains. RFdiffusion then generates a binder molecule that effectively connects the pockets into a single structure. Logos designed binders for 39 highly diverse unstructured targets and similarly obtained picomolar to nanomolar affinities in 34 cases.
Kejia Wu, PhD, a postdoctoral researcher in the Baker lab and co-lead author of the Science paper, has been working on disordered proteins since the beginning of her graduate school journey six years ago. She explained that the rise of powerful AI approaches, such as AlphaFold and RFdiffusion, has enabled an inflection point for protein design challenges that have historically faced a wall.
“I like this problem because it’s so messy. As scientists, we should be motivated to try and find the pattern from the mess,” Wu told GEN. “We’ve made a lot of progress from a computational protein design perspective and in in vitro studies.”
Baker emphasized that making binders to disordered proteins has “almost become easier” than ordered proteins because of the concept of induced fit. Given the unstructured nature of IDRs, both methods can sample a wide variety of conformations to identify top binding candidates. This notion contrasts with ordered protein counterparts, whose fixed structures restrict the number of optimal binding solutions.
“Even though the protein is unfolded on its own, it will adopt a specific structure when it interacts with the designed binder,” explained Baker. “The binder essentially forces the peptide into a conformation which is well suited to be bound.”
The Baker lab has made the complementary IDR binder design tools available online for all researchers to use. The next research steps will expand the methodology to account for IDR post-translational modifications involved in regulating biological function.
The post Undruggable No More: AI Hits Disordered Proteins, Unlocks Therapy Targets appeared first on GEN – Genetic Engineering and Biotechnology News.